The biorepository portal toolkit: an honest brokered, modular service oriented software tool set for biospecimen-driven translational research
- PMID: 27535360
- PMCID: PMC5001241
- DOI: 10.1186/s12864-016-2797-9
The biorepository portal toolkit: an honest brokered, modular service oriented software tool set for biospecimen-driven translational research
Abstract
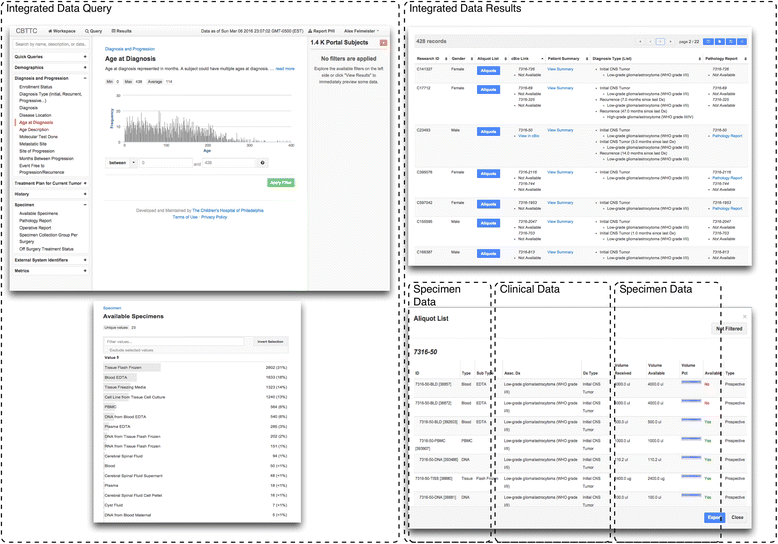
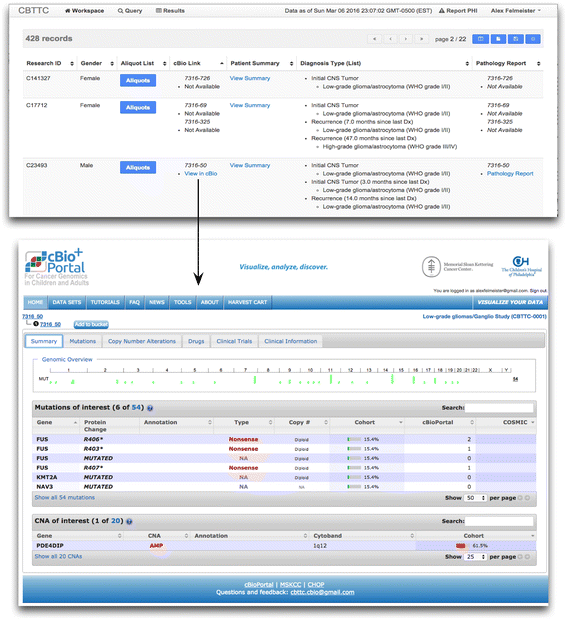
Background: High throughput molecular sequencing and increased biospecimen variety have introduced significant informatics challenges for research biorepository infrastructures. We applied a modular system integration approach to develop an operational biorepository management system. This method enables aggregation of the clinical, specimen and genomic data collected for biorepository resources.
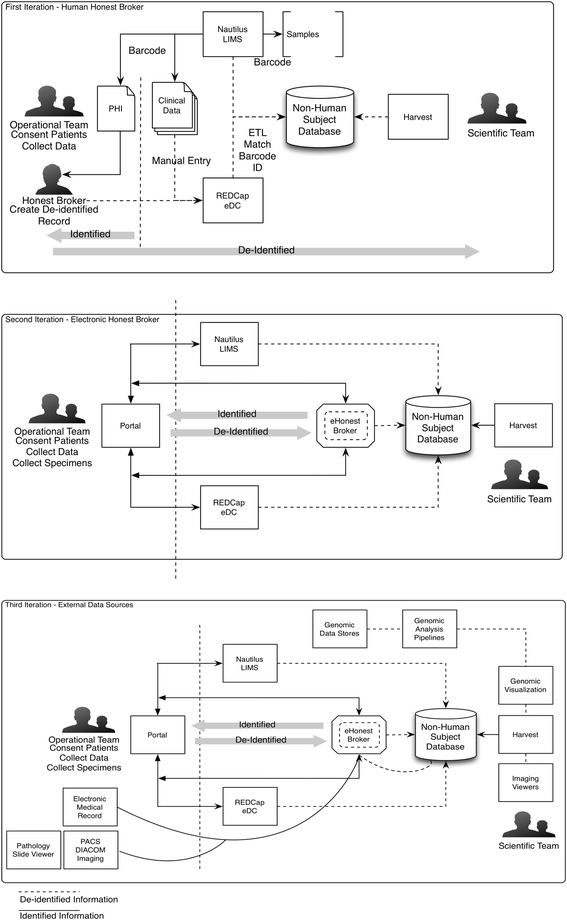
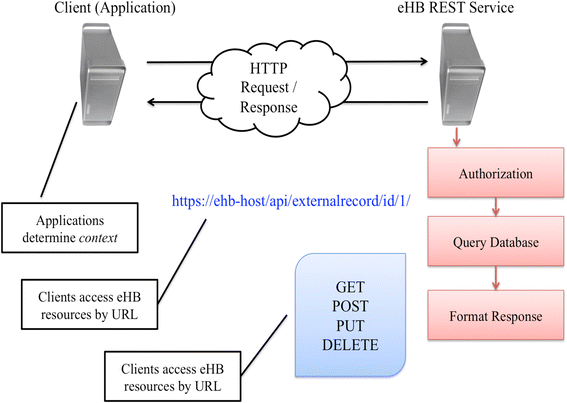
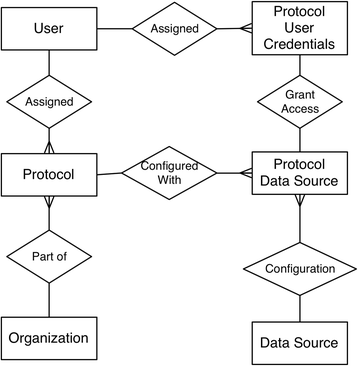
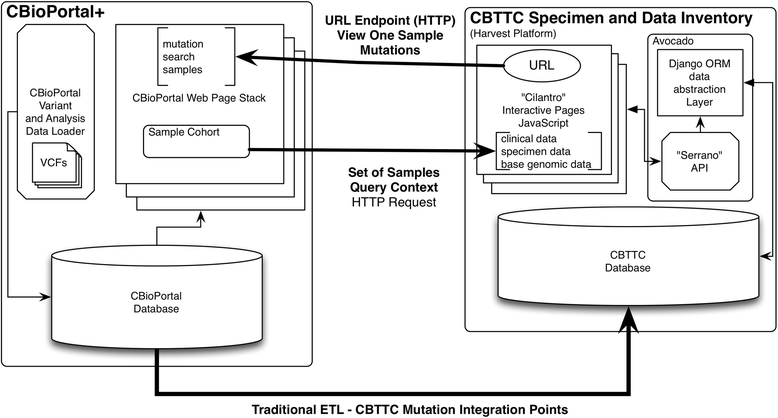
Methods: We introduce an electronic Honest Broker (eHB) and Biorepository Portal (BRP) open source project that, in tandem, allow for data integration while protecting patient privacy. This modular approach allows data and specimens to be associated with a biorepository subject at any time point asynchronously. This lowers the bar to develop new research projects based on scientific merit without institutional review for a proposal.
Results: By facilitating the automated de-identification of specimen and associated clinical and genomic data we create a future proofed specimen set that can withstand new workflows and be connected to new associated information over time. Thus facilitating collaborative advanced genomic and tissue research.
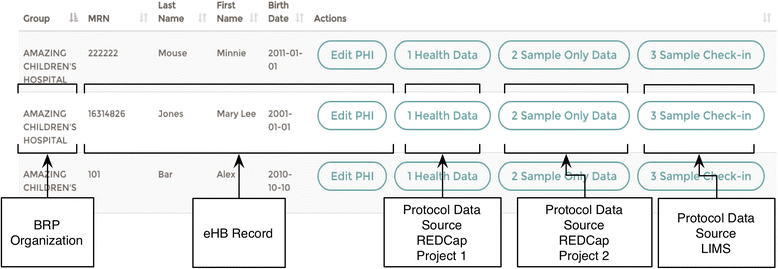
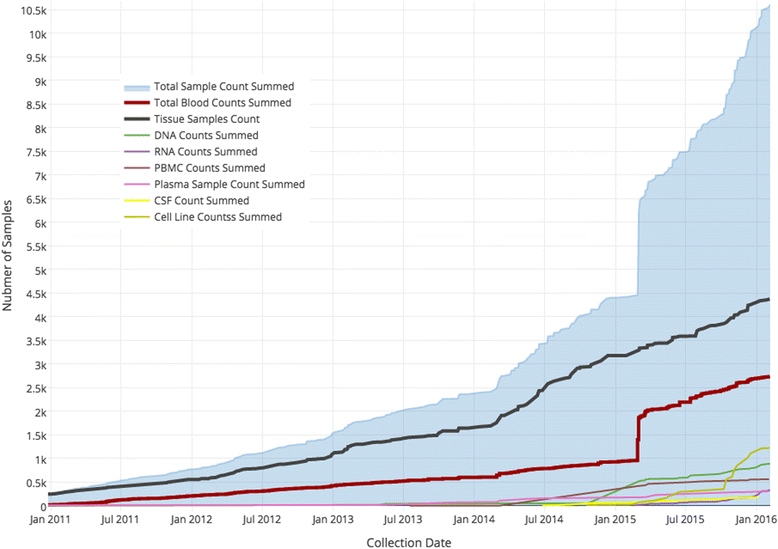
Conclusions: As of Janurary of 2016 there are 23 unique protocols/patient cohorts being managed in the Biorepository Portal (BRP). There are over 4000 unique subject records in the electronic honest broker (eHB), over 30,000 specimens accessioned and 8 institutions participating in various biobanking activities using this tool kit. We specifically set out to build rich annotation of biospecimens with longitudinal clinical data; BRP/REDCap integration for multi-institutional repositories; EMR integration; further annotated specimens with genomic data specific to a domain; build application hooks for experiments at the specimen level integrated with analytic software; while protecting privacy per the Office of Civil Rights (OCR) and HIPAA.
Keywords: Biorepository research; Cancer genomics; Data integration; Data representation; Honest broker; Open source; Patient health information protection; Patient privacy; Precision medicine; Translational bioinformatics.
Figures
Similar articles
-
REDCap and the National Mesothelioma Virtual Bank-a scalable and sustainable model for rare disease biorepositories.J Am Med Inform Assoc. 2023 Sep 25;30(10):1634-1644. doi: 10.1093/jamia/ocad132. J Am Med Inform Assoc. 2023. PMID: 37487555 Free PMC article.
-
A cervical cancer biorepository for pharmacogenomics research in Zimbabwe.BMC Cancer. 2022 Dec 16;22(1):1320. doi: 10.1186/s12885-022-10413-w. BMC Cancer. 2022. PMID: 36526993 Free PMC article.
-
caTissue Suite to OpenSpecimen: Developing an extensible, open source, web-based biobanking management system.J Biomed Inform. 2015 Oct;57:456-64. doi: 10.1016/j.jbi.2015.08.020. Epub 2015 Aug 29. J Biomed Inform. 2015. PMID: 26325296 Free PMC article.
-
Biorepositories and Databanks for the Development of Novel Biomarkers for Genitourinary Cancer Prevention and Management.Eur Urol Focus. 2021 May;7(3):513-521. doi: 10.1016/j.euf.2021.06.002. Epub 2021 Jun 22. Eur Urol Focus. 2021. PMID: 34167926 Review.
-
Biobanking Comes of Age: The Transition to Biospecimen Science.Annu Rev Pharmacol Toxicol. 2016;56:211-28. doi: 10.1146/annurev-pharmtox-010715-103246. Epub 2015 Oct 22. Annu Rev Pharmacol Toxicol. 2016. PMID: 26514206 Review.
Cited by
-
ARF suppression by MYC but not MYCN confers increased malignancy of aggressive pediatric brain tumors.Nat Commun. 2023 Mar 3;14(1):1221. doi: 10.1038/s41467-023-36847-9. Nat Commun. 2023. PMID: 36869047 Free PMC article.
-
Metabolomics technology and bioinformatics for precision medicine.Brief Bioinform. 2019 Nov 27;20(6):1957-1971. doi: 10.1093/bib/bbx170. Brief Bioinform. 2019. PMID: 29304189 Free PMC article. Review.
-
Multi-ancestry genome-wide association study of 4069 children with glioma identifies 9p21.3 risk locus.Neuro Oncol. 2023 Sep 5;25(9):1709-1720. doi: 10.1093/neuonc/noad042. Neuro Oncol. 2023. PMID: 36810956 Free PMC article.
-
Pro-inflammatory cytokines mediate the epithelial-to-mesenchymal-like transition of pediatric posterior fossa ependymoma.Nat Commun. 2022 Jul 8;13(1):3936. doi: 10.1038/s41467-022-31683-9. Nat Commun. 2022. PMID: 35803925 Free PMC article.
-
REDCap and the National Mesothelioma Virtual Bank-a scalable and sustainable model for rare disease biorepositories.J Am Med Inform Assoc. 2023 Sep 25;30(10):1634-1644. doi: 10.1093/jamia/ocad132. J Am Med Inform Assoc. 2023. PMID: 37487555 Free PMC article.
References
-
- Colman E, Golden J, Roberts M, Egan A, Weaver J, Pharm D, Rosebraugh C. The path to personalized medicine. N Engl J Med. 2010;363(4):2012–4.
-
- Hirtzlin I, Dubreuil C, Préaubert N, Duchier J, Jansen B, Simon J, Lobato De Faria P, Perez-Lezaun A, Visser B, Williams GD, Cambon-Thomsen A. An empirical survey on biobanking of human genetic material and data in six EU countries. Eur J Hum Genet. 2003;11(6):475–88. doi: 10.1038/sj.ejhg.5201007. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

