Analysis of protein-coding genetic variation in 60,706 humans
- PMID: 27535533
- PMCID: PMC5018207
- DOI: 10.1038/nature19057
Analysis of protein-coding genetic variation in 60,706 humans
Abstract
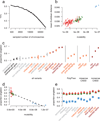
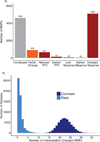
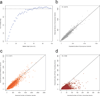
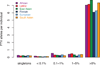
Large-scale reference data sets of human genetic variation are critical for the medical and functional interpretation of DNA sequence changes. Here we describe the aggregation and analysis of high-quality exome (protein-coding region) DNA sequence data for 60,706 individuals of diverse ancestries generated as part of the Exome Aggregation Consortium (ExAC). This catalogue of human genetic diversity contains an average of one variant every eight bases of the exome, and provides direct evidence for the presence of widespread mutational recurrence. We have used this catalogue to calculate objective metrics of pathogenicity for sequence variants, and to identify genes subject to strong selection against various classes of mutation; identifying 3,230 genes with near-complete depletion of predicted protein-truncating variants, with 72% of these genes having no currently established human disease phenotype. Finally, we demonstrate that these data can be used for the efficient filtering of candidate disease-causing variants, and for the discovery of human 'knockout' variants in protein-coding genes.
Figures


Comment in
-
Human genomics: A deep dive into genetic variation.Nature. 2016 Aug 18;536(7616):277-8. doi: 10.1038/536277a. Nature. 2016. PMID: 27535530 No abstract available.
-
Rethink the links between genes and disease.Nature. 2016 Oct 13;538(7624):140. doi: 10.1038/538140a. Nature. 2016. PMID: 27734882 No abstract available.
-
How scientists use Slack.Nature. 2016 Dec 29;541(7635):123-124. doi: 10.1038/541123a. Nature. 2016. PMID: 28054618 No abstract available.
References
-
- Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat. Rev. Genet. 2011;12:603–614. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54HG003067/HG/NHGRI NIH HHS/United States
- K02 NS085048/NS/NINDS NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- MOP82810/CAPMC/ CIHR/Canada
- RC2F DK088389/DK/NIDDK NIH HHS/United States
- U01-DK085545/DK/NIDDK NIH HHS/United States
- MH077139/MH/NIMH NIH HHS/United States
- HHSN268201300049C/HL/NHLBI NIH HHS/United States
- 098381/WT_/Wellcome Trust/United Kingdom
- U01 DK085545/DK/NIDDK NIH HHS/United States
- HHSN268201300046C/HL/NHLBI NIH HHS/United States
- NIMHRC2MH089905/PHS HHS/United States
- 1RC2DK088389/DK/NIDDK NIH HHS/United States
- 090367/WT_/Wellcome Trust/United Kingdom
- RG/13/13/30194/BHF_/British Heart Foundation/United Kingdom
- G0801418/MRC_/Medical Research Council/United Kingdom
- U01 DK085501/DK/NIDDK NIH HHS/United States
- 2P50MH066392-05A1/MH/NIMH NIH HHS/United States
- R01 MH077139/MH/NIMH NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- MH095034/MH/NIMH NIH HHS/United States
- MOP136936/CAPMC/ CIHR/Canada
- R01HL107816/HL/NHLBI NIH HHS/United States
- R01 DK098032/DK/NIDDK NIH HHS/United States
- U01DK085526/DK/NIDDK NIH HHS/United States
- U01 NS040024/NS/NINDS NIH HHS/United States
- HHSN268201300047C/HL/NHLBI NIH HHS/United States
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- R01DK062370/DK/NIDDK NIH HHS/United States
- U41 HG000330/HG/NHGRI NIH HHS/United States
- K01 HL125751/HL/NHLBI NIH HHS/United States
- T32 HL007208/HL/NHLBI NIH HHS/United States
- G0800509/MRC_/Medical Research Council/United Kingdom
- U01 DK085584/DK/NIDDK NIH HHS/United States
- MOP77682/CAPMC/ CIHR/Canada
- HHSN268201300048C/HL/NHLBI NIH HHS/United States
- U01 DK085524/DK/NIDDK NIH HHS/United States
- MC_UP_1102/20/MRC_/Medical Research Council/United Kingdom
- 5U54HG003067-11/HG/NHGRI NIH HHS/United States
- R01DK098032/DK/NIDDK NIH HHS/United States
- RC2DK088389/DK/NIDDK NIH HHS/United States
- DK085545/DK/NIDDK NIH HHS/United States
- U01 DK085526/DK/NIDDK NIH HHS/United States
- R01MH085521/MH/NIMH NIH HHS/United States
- MH094421/MH/NIMH NIH HHS/United States
- NS40024-09S1/NS/NINDS NIH HHS/United States
- DK088389/DK/NIDDK NIH HHS/United States
- DK098032/DK/NIDDK NIH HHS/United States
- U01 DK062370/DK/NIDDK NIH HHS/United States
- P30 AG038072/AG/NIA NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- U01 NS40024-09S1/NS/NINDS NIH HHS/United States
- RC2-DK088389/DK/NIDDK NIH HHS/United States
- FS/14/55/30806/BHF_/British Heart Foundation/United Kingdom
- R01HL24799/HL/NHLBI NIH HHS/United States
- U54 DK105566/DK/NIDDK NIH HHS/United States
- 5 U54 HG003067-13/HG/NHGRI NIH HHS/United States
- U01 MH094432/MH/NIMH NIH HHS/United States
- R01 GM104371/GM/NIGMS NIH HHS/United States
- HHSN268201300050C/HL/NHLBI NIH HHS/United States
- K01HL125751/HL/NHLBI NIH HHS/United States
- F32GM115208/GM/NIGMS NIH HHS/United States
- MH089905/MH/NIMH NIH HHS/United States
- R01MH085560/MH/NIMH NIH HHS/United States
- NS085048/NS/NINDS NIH HHS/United States
- G0601261/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

