A new transcriptome and transcriptome profiling of adult and larval tissue in the box jellyfish Alatina alata: an emerging model for studying venom, vision and sex
- PMID: 27535656
- PMCID: PMC4989536
- DOI: 10.1186/s12864-016-2944-3
A new transcriptome and transcriptome profiling of adult and larval tissue in the box jellyfish Alatina alata: an emerging model for studying venom, vision and sex
Erratum in
-
Erratum to: A new transcriptome and transcriptome profiling of adult and larval tissue in the box jellyfish Alatina alata: an emerging model for studying venom, vision and sex.BMC Genomics. 2016 Nov 28;17(1):980. doi: 10.1186/s12864-016-3305-y. BMC Genomics. 2016. PMID: 27894263 Free PMC article. No abstract available.
Abstract
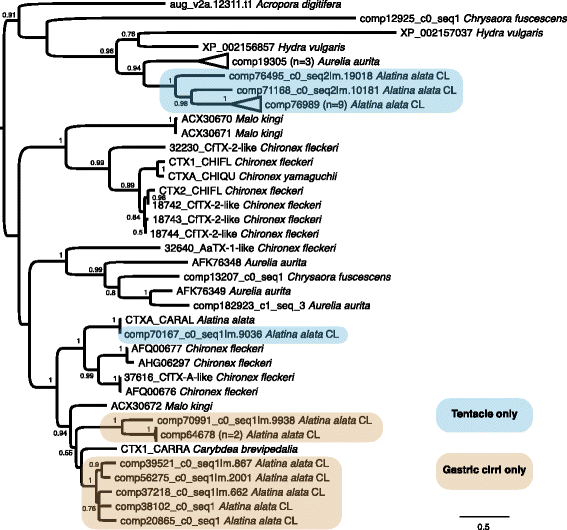
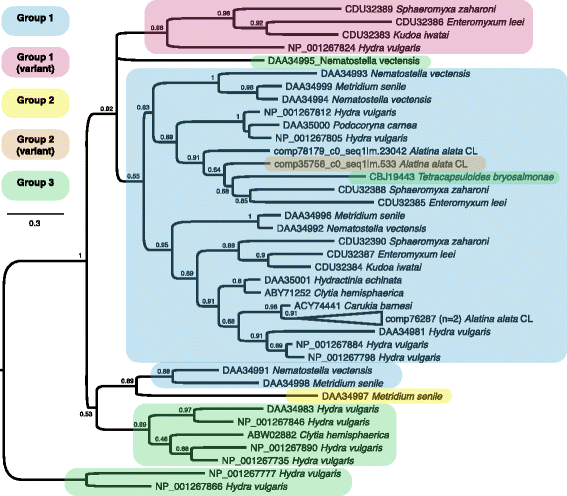
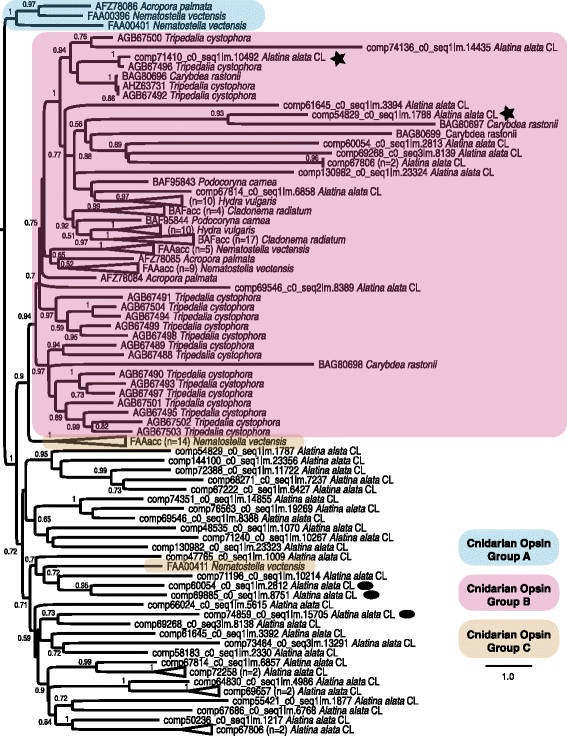
Background: Cubozoans (box jellyfish) are cnidarians that have evolved a number of distinguishing features. Many cubozoans have a particularly potent sting, effected by stinging structures called nematocysts; cubozoans have well-developed light sensation, possessing both image-forming lens eyes and light-sensitive eye spots; and some cubozoans have complex mating behaviors, including aggregations, copulation and internal fertilization. The cubozoan Alatina alata is emerging as a cnidarian model because it forms predictable monthly nearshore breeding aggregations in tropical to subtropical waters worldwide, making both adult and larval material reliably accessible. To develop resources for A. alata, this study generated a functionally annotated transcriptome of adult and larval tissue, applying preliminary differential expression analyses to identify candidate genes involved in nematogenesis and venom production, vision and extraocular sensory perception, and sexual reproduction, which for brevity we refer to as "venom", "vision" and "sex".
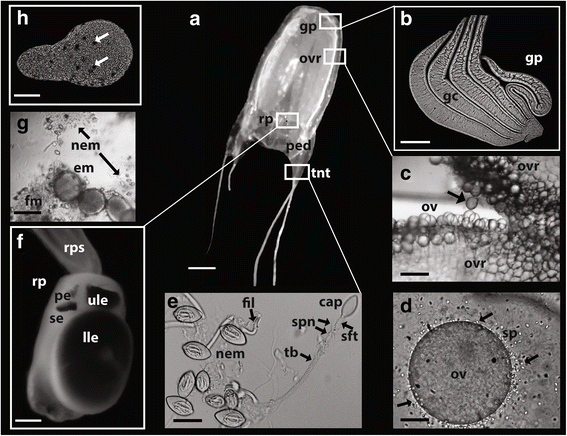
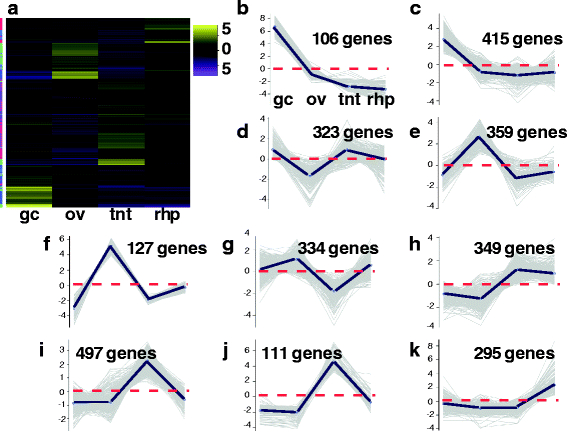
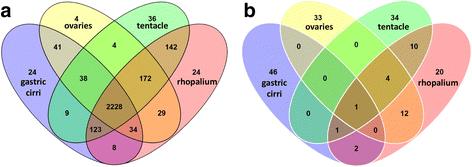
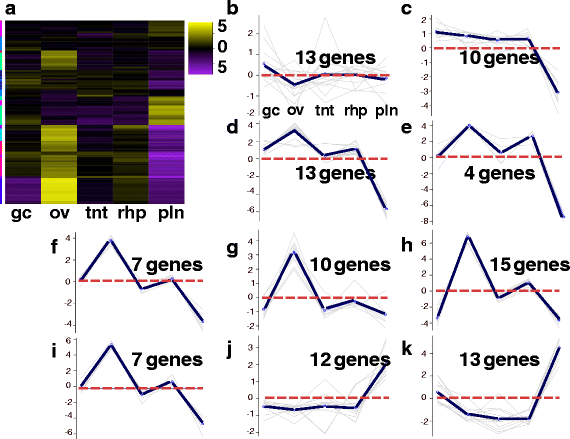
Results: We assembled a transcriptome de novo from RNA-Seq data pooled from multiple body parts (gastric cirri, ovaries, tentacle (with pedalium base) and rhopalium) of an adult female A. alata medusa and larval planulae. Our transcriptome comprises ~32 K transcripts, after filtering, and provides a basis for analyzing patterns of gene expression in adult and larval box jellyfish tissues. Furthermore, we annotated a large set of candidate genes putatively involved in venom, vision and sex, providing an initial molecular characterization of these complex features in cubozoans. Expression profiles and gene tree reconstruction provided a number of preliminary insights into the putative sites of nematogenesis and venom production, regions of phototransduction activity and fertilization dynamics in A. alata.
Conclusions: Our Alatina alata transcriptome significantly adds to the genomic resources for this emerging cubozoan model. This study provides the first annotated transcriptome from multiple tissues of a cubozoan focusing on both the adult and larvae. Our approach of using multiple body parts and life stages to generate this transcriptome effectively identified a broad range of candidate genes for the further study of coordinated processes associated with venom, vision and sex. This new genomic resource and the candidate gene dataset are valuable for further investigating the evolution of distinctive features of cubozoans, and of cnidarians more broadly.
Keywords: Cubozoa; Embryo; Expression patterns; Eye; Gametogenesis; Pedalium; Planulae; Spawning aggregations; Sperm; Sting.
Figures





References
-
- Lewis C, Long TAF. Courtship and reproduction in Carybdea sivickisi (Cnidaria: Cubozoa). Mar. Biol. [Internet]. 2005 [cited 2014 Feb 27];147:477–83. Available from: http://link.springer.com/10.1007/s00227-005-1602-0 - DOI
-
- Garm A, Lebouvier M, Tolunay D. Mating in the box jellyfish Copula sivickisi-Novel function of cnidocytes. J. Morphol. [Internet]. 2015 [cited 2015 Jun 4];00. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26010863 - PubMed
-
- Lewis C, Bentlage B. Clarifying the identity of the Japanese Habu-kurage, Chironex yamaguchii, sp. nov. (Cnidaria: Cubozoa: Chirodropida) Zootaxa. 2009;65:59–65.
-
- Brinkman DL, Jia X, Potriquet J, Kumar D, Dash D, Kvaskoff D, et al. Transcriptome and venom proteome of the box jellyfish Chironex fleckeri. BMC Genomics [Internet]. 2015 [cited 2015 Jun 5];16:407. Available from: http://www.biomedcentral.com/1471-2164/16/407 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

