Pangenome Evidence for Higher Codon Usage Bias and Stronger Translational Selection in Core Genes of Escherichia coli
- PMID: 27536275
- PMCID: PMC4971109
- DOI: 10.3389/fmicb.2016.01180
Pangenome Evidence for Higher Codon Usage Bias and Stronger Translational Selection in Core Genes of Escherichia coli
Abstract
Codon usage bias, as a combined interplay from mutation and selection, has been intensively studied in Escherichia coli. However, codon usage analysis in an E. coli pangenome remains unexplored and the relative importance of mutation and selection acting on core genes and strain-specific genes is unknown. Here we perform comprehensive codon usage analyses based on a collection of multiple complete genome sequences of E. coli. Our results show that core genes that are present in all strains have higher codon usage bias than strain-specific genes that are unique to single strains. We further explore the forces in influencing codon usage and investigate the difference of the major force between core and strain-specific genes. Our results demonstrate that although mutation may exert genome-wide influences on codon usage acting similarly in different gene sets, selection dominates as an important force to shape biased codon usage as genes are present in an increased number of strains. Together, our results provide important insights for better understanding genome plasticity and complexity as well as evolutionary mechanisms behind codon usage bias.
Keywords: codon usage bias; core genes; mutation; pangenome; strain-specific genes; translational selection.
Figures

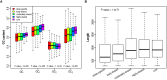
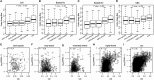
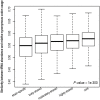

Similar articles
-
Analysis of synonymous codon usage in Shigella flexneri 2a strain 301 and other Shigella and Escherichia coli strains.Can J Microbiol. 2011 Dec;57(12):1016-23. doi: 10.1139/w11-095. Epub 2011 Nov 23. Can J Microbiol. 2011. PMID: 22112197
-
Codon usage in highly expressed genes of Haemophillus influenzae and Mycobacterium tuberculosis: translational selection versus mutational bias.Gene. 1998 Jul 30;215(2):405-13. doi: 10.1016/s0378-1119(98)00257-1. Gene. 1998. PMID: 9714839
-
Codon Usage Heterogeneity in the Multipartite Prokaryote Genome: Selection-Based Coding Bias Associated with Gene Location, Expression Level, and Ancestry.mBio. 2019 May 28;10(3):e00505-19. doi: 10.1128/mBio.00505-19. mBio. 2019. PMID: 31138741 Free PMC article.
-
Estimating Gene Expression and Codon-Specific Translational Efficiencies, Mutation Biases, and Selection Coefficients from Genomic Data Alone.Genome Biol Evol. 2015 May 14;7(6):1559-79. doi: 10.1093/gbe/evv087. Genome Biol Evol. 2015. PMID: 25977456 Free PMC article.
-
Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes.Biol Rev Camb Philos Soc. 2013 Feb;88(1):49-61. doi: 10.1111/j.1469-185X.2012.00242.x. Epub 2012 Aug 14. Biol Rev Camb Philos Soc. 2013. PMID: 22889422 Review.
Cited by
-
Comparative Genomic Analysis of Soil Dwelling Bacteria Utilizing a Combinational Codon Usage and Molecular Phylogenetic Approach Accentuating on Key Housekeeping Genes.Front Microbiol. 2019 Dec 17;10:2896. doi: 10.3389/fmicb.2019.02896. eCollection 2019. Front Microbiol. 2019. PMID: 31921071 Free PMC article.
-
Synonymous sites for accessibility around microRNA binding sites in bacterial spot and speck disease resistance genes of tomato.Funct Integr Genomics. 2023 Jul 20;23(3):247. doi: 10.1007/s10142-023-01178-x. Funct Integr Genomics. 2023. PMID: 37468805
-
A Study on microRNAs Targeting the Genes Overexpressed in Lung Cancer and their Codon Usage Patterns.Mol Biotechnol. 2022 Oct;64(10):1095-1119. doi: 10.1007/s12033-022-00491-3. Epub 2022 Apr 18. Mol Biotechnol. 2022. PMID: 35435592
-
The genomic configurations driving antimicrobial resistance and virulence in colistin resistant Pseudomonas aeruginosa from an Egyptian Tertiary Oncology Hospital.PLOS Glob Public Health. 2025 Aug 5;5(8):e0004976. doi: 10.1371/journal.pgph.0004976. eCollection 2025. PLOS Glob Public Health. 2025. PMID: 40763289 Free PMC article.
-
DNA Segregation in Enterobacteria.EcoSal Plus. 2023 Dec 12;11(1):eesp00382020. doi: 10.1128/ecosalplus.esp-0038-2020. Epub 2023 May 9. EcoSal Plus. 2023. PMID: 37220081 Free PMC article. Review.
References
-
- Blattner F. R., Plunkett G., Bloch C. A., Perna N. T., Burland V., Riley M., et al. . (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

