Heterokont Predator Develorapax marinus gen. et sp. nov. - A Model of the Ochrophyte Ancestor
- PMID: 27536283
- PMCID: PMC4971089
- DOI: 10.3389/fmicb.2016.01194
Heterokont Predator Develorapax marinus gen. et sp. nov. - A Model of the Ochrophyte Ancestor
Abstract
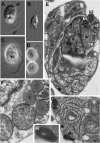
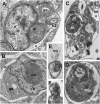
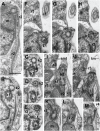
Heterotrophic lineages of Heterokonta (or stramenopiles), in contrast to a single monophyletic group of autotrophs, Ochrophyta, form several clades that independently branch off the heterokont stem lineage. The nearest neighbors of Ochrophyta in the phylogenetic tree appear to be almost exclusively bacterivorous, whereas the hypothesis of plastid acquisition by the ancestors of the ochrophyte lineage suggests an ability to engulf eukaryotic alga. In line with this hypothesis, the heterotrophic predator at the base of the ochrophyte lineage may be regarded as a model for the ochrophyte ancestor. Here, we present a new genus and species of marine free-living heterotrophic heterokont Develorapax marinus, which falls into an isolated heterokont cluster, along with the marine flagellate Developayella elegans, and is able to engulf eukaryotic cells. Together with environmental sequences D. marinus and D. elegans form a class-level clade Developea nom. nov. represented by species adapted to different environmental conditions and with a wide geographical distribution. The position of Developea among Heterokonta in large-scale phylogenetic tree is discussed. We propose that members of the Developea clade represent a model for transition from bacterivory to a predatory feeding mode by selection for larger prey. Presumably, such transition in the grazing strategy is possible in the presence of bacterial biofilms or aggregates expected in eutrophic environment, and has likely occurred in the ochrophyte ancestor.
Keywords: Develorapax marinus; molecular phylogeny; ochrophyte ancestor; prey size; ultrastructure.
Figures

References
-
- Al-Qassab S., Lee W. J., Murray S., Simpson A. G. B., Patterson D. J. (2002). Flagellates from stromatolites and surrounding sediments in Shark Bay, Western Australia. Acta Protozool. 41 91–144.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

