Proteomic Response of Hordeum vulgare cv. Tadmor and Hordeum marinum to Salinity Stress: Similarities and Differences between a Glycophyte and a Halophyte
- PMID: 27536311
- PMCID: PMC4971088
- DOI: 10.3389/fpls.2016.01154
Proteomic Response of Hordeum vulgare cv. Tadmor and Hordeum marinum to Salinity Stress: Similarities and Differences between a Glycophyte and a Halophyte
Abstract
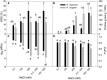
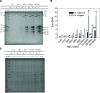
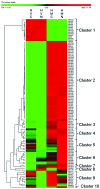
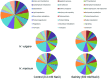
Response to a high salinity treatment of 300 mM NaCl was studied in a cultivated barley Hordeum vulgare Syrian cultivar Tadmor and in a halophytic wild barley H. marinum. Differential salinity tolerance of H. marinum and H. vulgare is underlied by qualitative and quantitative differences in proteins involved in a variety of biological processes. The major aim was to identify proteins underlying differential salinity tolerance between the two barley species. Analyses of plant water content, osmotic potential and accumulation of proline and dehydrin proteins under high salinity revealed a relatively higher water saturation deficit in H. marinum than in H. vulgare while H. vulgare had lower osmotic potential corresponding with high levels of proline and dehydrins. Analysis of proteins soluble upon boiling isolated from control and salt-treated crown tissues revealed similarities as well as differences between H. marinum and H. vulgare. The similar salinity responses of both barley species lie in enhanced levels of stress-protective proteins such as defense-related proteins from late-embryogenesis abundant family, several chaperones from heat shock protein family, and others such as GrpE. However, there have also been found significant differences between H. marinum and H. vulgare salinity response indicating an active stress acclimation in H. marinum while stress damage in H. vulgare. An active acclimation to high salinity in H. marinum is underlined by enhanced levels of several stress-responsive transcription factors from basic leucine zipper and nascent polypeptide-associated complex families. In salt-treated H. marinum, enhanced levels of proteins involved in energy metabolism such as glycolysis, ATP metabolism, and photosynthesis-related proteins indicate an active acclimation to enhanced energy requirements during an establishment of novel plant homeostasis. In contrast, changes at proteome level in salt-treated H. vulgare indicate plant tissue damage as revealed by enhanced levels of proteins involved in proteasome-dependent protein degradation and proteins related to apoptosis. The results of proteomic analysis clearly indicate differential responses to high salinity and provide more profound insight into biological mechanisms underlying salinity response between two barley species with contrasting salinity tolerance.
Keywords: Hordeum marinum; Hordeum vulgare; glycophyte; halophyte; proteome; salinity; stress acclimation; stress damage.
Figures


Similar articles
-
Adaptation Strategies of Halophytic Barley Hordeum marinum ssp. marinum to High Salinity and Osmotic Stress.Int J Mol Sci. 2020 Nov 27;21(23):9019. doi: 10.3390/ijms21239019. Int J Mol Sci. 2020. PMID: 33260985 Free PMC article.
-
Differences in efficient metabolite management and nutrient metabolic regulation between wild and cultivated barley grown at high salinity.Plant Biol (Stuttg). 2010 Jul 1;12(4):650-8. doi: 10.1111/j.1438-8677.2009.00265.x. Plant Biol (Stuttg). 2010. PMID: 20636908
-
Identification of proteins associated with ion homeostasis and salt tolerance in barley.Proteomics. 2014 Jun;14(11):1381-92. doi: 10.1002/pmic.201300221. Epub 2014 Apr 16. Proteomics. 2014. PMID: 24616274
-
Wheat and barley dehydrins under cold, drought, and salinity - what can LEA-II proteins tell us about plant stress response?Front Plant Sci. 2014 Jul 9;5:343. doi: 10.3389/fpls.2014.00343. eCollection 2014. Front Plant Sci. 2014. PMID: 25071816 Free PMC article. Review.
-
Proteomics of stress responses in wheat and barley-search for potential protein markers of stress tolerance.Front Plant Sci. 2014 Dec 11;5:711. doi: 10.3389/fpls.2014.00711. eCollection 2014. Front Plant Sci. 2014. PMID: 25566285 Free PMC article. Review.
Cited by
-
Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome.Front Plant Sci. 2018 Feb 8;9:122. doi: 10.3389/fpls.2018.00122. eCollection 2018. Front Plant Sci. 2018. PMID: 29472941 Free PMC article. Review.
-
Hypusination, a Metabolic Posttranslational Modification of eIF5A in Plants during Development and Environmental Stress Responses.Plants (Basel). 2021 Jun 22;10(7):1261. doi: 10.3390/plants10071261. Plants (Basel). 2021. PMID: 34206171 Free PMC article. Review.
-
Morphological Analysis, Protein Profiling and Expression Analysis of Auxin Homeostasis Genes of Roots of Two Contrasting Cultivars of Rice Provide Inputs on Mechanisms Involved in Rice Adaptation towards Salinity Stress.Plants (Basel). 2021 Jul 28;10(8):1544. doi: 10.3390/plants10081544. Plants (Basel). 2021. PMID: 34451587 Free PMC article.
-
ROS scavenging and ion homeostasis is required for the adaptation of halophyte Karelinia caspia to high salinity.Front Plant Sci. 2022 Oct 3;13:979956. doi: 10.3389/fpls.2022.979956. eCollection 2022. Front Plant Sci. 2022. PMID: 36262663 Free PMC article.
-
Magnesium Deficiency Induced Global Transcriptome Change in Citrus sinensis Leaves Revealed by RNA-Seq.Int J Mol Sci. 2019 Jun 26;20(13):3129. doi: 10.3390/ijms20133129. Int J Mol Sci. 2019. PMID: 31248059 Free PMC article.
References
-
- Athar H. R., Zafar Z. U., Ashraf M. (2015). Glycinebetaine improved photosynthesis in canola under salt stress: evaluation of chlorophyll fluorescence parameters as potential indicators. J. Agron. Crop Sci. 201 428–442. 10.1111/jac.12120 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

