Systematically Altering Bacterial SOS Activity under Stress Reveals Therapeutic Strategies for Potentiating Antibiotics
- PMID: 27536734
- PMCID: PMC4980697
- DOI: 10.1128/mSphere.00163-16
Systematically Altering Bacterial SOS Activity under Stress Reveals Therapeutic Strategies for Potentiating Antibiotics
Abstract
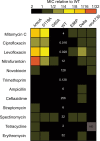
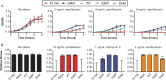
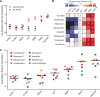
The bacterial SOS response is a DNA damage repair network that is strongly implicated in both survival and acquired drug resistance under antimicrobial stress. The two SOS regulators, LexA and RecA, have therefore emerged as potential targets for adjuvant therapies aimed at combating resistance, although many open questions remain. For example, it is not well understood whether SOS hyperactivation is a viable therapeutic approach or whether LexA or RecA is a better target. Furthermore, it is important to determine which antimicrobials could serve as the best treatment partners with SOS-targeting adjuvants. Here we derived Escherichia coli strains that have mutations in either lexA or recA genes in order to cover the full spectrum of possible SOS activity levels. We then systematically analyzed a wide range of antimicrobials by comparing the mean inhibitory concentrations (MICs) and induced mutation rates for each drug-strain combination. We first show that significant changes in MICs are largely confined to DNA-damaging antibiotics, with strains containing a constitutively repressed SOS response impacted to a greater extent than hyperactivated strains. Second, antibiotic-induced mutation rates were suppressed when SOS activity was reduced, and this trend was observed across a wider spectrum of antibiotics. Finally, perturbing either LexA or RecA proved to be equally viable strategies for targeting the SOS response. Our work provides support for multiple adjuvant strategies, while also suggesting that the combination of an SOS inhibitor with a DNA-damaging antibiotic could offer the best potential for lowering MICs and decreasing acquired drug resistance. IMPORTANCE Our antibiotic arsenal is becoming depleted, in part, because bacteria have the ability to rapidly adapt and acquire resistance to our best agents. The SOS pathway, a widely conserved DNA damage stress response in bacteria, is activated by many antibiotics and has been shown to play central role in promoting survival and the evolution of resistance under antibiotic stress. As a result, targeting the SOS response has been proposed as an adjuvant strategy to revitalize our current antibiotic arsenal. However, the optimal molecular targets and partner antibiotics for such an approach remain unclear. In this study, focusing on the two key regulators of the SOS response, LexA and RecA, we provide the first comprehensive assessment of how to target the SOS response in order to increase bacterial susceptibility and reduce mutagenesis under antibiotic treatment.
Keywords: DNA damage; LexA; RecA; SOS pathway; adjuvant therapy; antibiotic resistance; mutagenesis.
Figures

References
-
- Centers for Disease Control and Prevention 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA. http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
-
- Centers for Disease Control and Prevention 2014. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2012. Centers for Disease Control and Prevention, U. S. Department of Health and Human Services, Atlanta, GA: http://www.cdc.gov/narms/pdf/2012-annual-report-narms-508c.pdf.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
