Intercellular Variability in Protein Levels from Stochastic Expression and Noisy Cell Cycle Processes
- PMID: 27536771
- PMCID: PMC4990281
- DOI: 10.1371/journal.pcbi.1004972
Intercellular Variability in Protein Levels from Stochastic Expression and Noisy Cell Cycle Processes
Abstract
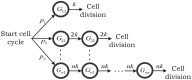
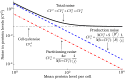
Inside individual cells, expression of genes is inherently stochastic and manifests as cell-to-cell variability or noise in protein copy numbers. Since proteins half-lives can be comparable to the cell-cycle length, randomness in cell-division times generates additional intercellular variability in protein levels. Moreover, as many mRNA/protein species are expressed at low-copy numbers, errors incurred in partitioning of molecules between two daughter cells are significant. We derive analytical formulas for the total noise in protein levels when the cell-cycle duration follows a general class of probability distributions. Using a novel hybrid approach the total noise is decomposed into components arising from i) stochastic expression; ii) partitioning errors at the time of cell division and iii) random cell-division events. These formulas reveal that random cell-division times not only generate additional extrinsic noise, but also critically affect the mean protein copy numbers and intrinsic noise components. Counter intuitively, in some parameter regimes, noise in protein levels can decrease as cell-division times become more stochastic. Computations are extended to consider genome duplication, where transcription rate is increased at a random point in the cell cycle. We systematically investigate how the timing of genome duplication influences different protein noise components. Intriguingly, results show that noise contribution from stochastic expression is minimized at an optimal genome-duplication time. Our theoretical results motivate new experimental methods for decomposing protein noise levels from synchronized and asynchronized single-cell expression data. Characterizing the contributions of individual noise mechanisms will lead to precise estimates of gene expression parameters and techniques for altering stochasticity to change phenotype of individual cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Quantifying intrinsic and extrinsic variability in stochastic gene expression models.PLoS One. 2013 Dec 31;8(12):e84301. doi: 10.1371/journal.pone.0084301. eCollection 2013. PLoS One. 2013. PMID: 24391934 Free PMC article.
-
Effects of cell-cycle-dependent expression on random fluctuations in protein levels.R Soc Open Sci. 2016 Dec 7;3(12):160578. doi: 10.1098/rsos.160578. eCollection 2016 Dec. R Soc Open Sci. 2016. PMID: 28083102 Free PMC article.
-
Interplay between intrinsic noise and the stochasticity of the cell cycle in bacterial colonies.Biophys J. 2010 Jun 2;98(11):2459-68. doi: 10.1016/j.bpj.2010.02.045. Biophys J. 2010. PMID: 20513389 Free PMC article.
-
Stochastic models coupling gene expression and partitioning in cell division in Escherichia coli.Biosystems. 2020 Jun;193-194:104154. doi: 10.1016/j.biosystems.2020.104154. Epub 2020 Apr 28. Biosystems. 2020. PMID: 32353481 Review.
-
Modeling cell-to-cell stochastic variability in intrinsic apoptosis pathway.Annu Int Conf IEEE Eng Med Biol Soc. 2012;2012:5498-501. doi: 10.1109/EMBC.2012.6347239. Annu Int Conf IEEE Eng Med Biol Soc. 2012. PMID: 23367174 Review.
Cited by
-
Collective behavior and self-organization in neural rosette morphogenesis.Front Cell Dev Biol. 2023 Aug 10;11:1134091. doi: 10.3389/fcell.2023.1134091. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37635866 Free PMC article. Review.
-
A mechanism for epithelial-mesenchymal heterogeneity in a population of cancer cells.PLoS Comput Biol. 2020 Feb 10;16(2):e1007619. doi: 10.1371/journal.pcbi.1007619. eCollection 2020 Feb. PLoS Comput Biol. 2020. PMID: 32040502 Free PMC article.
-
Quantitative measurement of phenotype dynamics during cancer drug resistance evolution using genetic barcoding.Nat Commun. 2025 Jun 20;16(1):5282. doi: 10.1038/s41467-025-59479-7. Nat Commun. 2025. PMID: 40541962 Free PMC article.
-
Terminal Schwann cell and vacant site mediated synapse elimination at developing neuromuscular junctions.Sci Rep. 2019 Dec 9;9(1):18594. doi: 10.1038/s41598-019-55017-w. Sci Rep. 2019. PMID: 31819113 Free PMC article.
-
Intrinsic and extrinsic noise of gene expression in lineage trees.Sci Rep. 2019 Jan 24;9(1):474. doi: 10.1038/s41598-018-35927-x. Sci Rep. 2019. PMID: 30679440 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

