Molecular Models of STAT5A Tetramers Complexed to DNA Predict Relative Genome-Wide Frequencies of the Spacing between the Two Dimer Binding Motifs of the Tetramer Binding Sites
- PMID: 27537504
- PMCID: PMC4990345
- DOI: 10.1371/journal.pone.0160339
Molecular Models of STAT5A Tetramers Complexed to DNA Predict Relative Genome-Wide Frequencies of the Spacing between the Two Dimer Binding Motifs of the Tetramer Binding Sites
Abstract
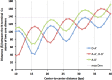
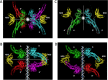
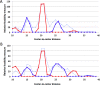
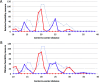
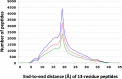
STAT proteins bind DNA as dimers and tetramers to control cellular development, differentiation, survival, and expansion. The tetramer binding sites are comprised of two dimer-binding sites repeated in tandem. The genome-wide distribution of the spacings between the dimer binding sites shows a distinctive, non-random pattern. Here, we report on estimating the feasibility of building possible molecular models of STAT5A tetramers bound to a DNA double helix with all possible spacings between the dimer binding sites. We found that the calculated feasibility estimates correlated well with the experimentally measured frequency of tetramer-binding sites. This suggests that the feasibility of forming the tetramer complex was a major factor in the evolution of this DNA sequence variation.
Conflict of interest statement
Figures

Similar articles
-
DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites.Mol Cell Biol. 2000 Jan;20(1):389-401. doi: 10.1128/MCB.20.1.389-401.2000. Mol Cell Biol. 2000. PMID: 10594041 Free PMC article.
-
Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function.Immunity. 2012 Apr 20;36(4):586-99. doi: 10.1016/j.immuni.2012.02.017. Immunity. 2012. PMID: 22520852 Free PMC article.
-
Stat5 tetramer formation is associated with leukemogenesis.Cancer Cell. 2005 Jan;7(1):87-99. doi: 10.1016/j.ccr.2004.12.010. Cancer Cell. 2005. PMID: 15652752
-
How p53 binds DNA as a tetramer.EMBO J. 1998 Jun 15;17(12):3342-50. doi: 10.1093/emboj/17.12.3342. EMBO J. 1998. PMID: 9628871 Free PMC article.
-
Structure of the unphosphorylated STAT5a dimer.J Biol Chem. 2005 Dec 9;280(49):40782-7. doi: 10.1074/jbc.M507682200. Epub 2005 Sep 28. J Biol Chem. 2005. PMID: 16192273
Cited by
-
Identification of mutations in porcine STAT5A that contributes to the transcription of CISH.Front Vet Sci. 2023 Jan 17;9:1090833. doi: 10.3389/fvets.2022.1090833. eCollection 2022. Front Vet Sci. 2023. PMID: 36733428 Free PMC article.
-
Fine-Tuning Cytokine Signals.Annu Rev Immunol. 2019 Apr 26;37:295-324. doi: 10.1146/annurev-immunol-042718-041447. Epub 2019 Jan 16. Annu Rev Immunol. 2019. PMID: 30649989 Free PMC article. Review.
-
STAT5 as a Key Protein of Erythropoietin Signalization.Int J Mol Sci. 2021 Jul 1;22(13):7109. doi: 10.3390/ijms22137109. Int J Mol Sci. 2021. PMID: 34281163 Free PMC article. Review.
-
Direct targets of pSTAT5 signalling in erythropoiesis.PLoS One. 2017 Jul 21;12(7):e0180922. doi: 10.1371/journal.pone.0180922. eCollection 2017. PLoS One. 2017. PMID: 28732065 Free PMC article.
-
STAT5A induced LINC01198 promotes proliferation of glioma cells through stabilizing DGCR8.Aging (Albany NY). 2020 Apr 4;12(7):5675-5692. doi: 10.18632/aging.102938. Epub 2020 Apr 4. Aging (Albany NY). 2020. PMID: 32246817 Free PMC article.
References
-
- Leonard WJ, O'Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. Epub 1998/05/23. 10.1146/annurev.immunol.16.1.293. . - PubMed
-
- Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, et al. Stat5 tetramer formation is associated with leukemogenesis. Cancer cell. 2005;7(1):87–99. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

