Notch-Hes-1 axis controls TLR7-mediated autophagic death of macrophage via induction of P62 in mice with lupus
- PMID: 27537524
- PMCID: PMC5108329
- DOI: 10.1038/cddis.2016.244
Notch-Hes-1 axis controls TLR7-mediated autophagic death of macrophage via induction of P62 in mice with lupus
Abstract
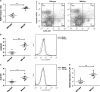
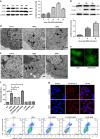
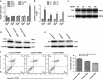
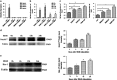
The increased death of macrophages has been considered as a pathogenic factor for systemic lupus erythematosus (SLE), and dysfunction of autophagy may contribute to improper cell death. However, the effect of autophagy on macrophage during the pathogenesis of SLE is still unclear. Here we found that the death rate and autophagy level of macrophages significantly increased in MRL/lpr lupus-prone mice. Activation of toll-like receptor 7 (TLR7) triggered macrophage death in an autophagy-dependent but caspase-independent way in vitro. Moreover, P62/SQSTM1 is thought to have an essential role in selective autophagy. We also demonstrated that P62/SQSTM1 was required for TLR7-induced autophagy, and knockdown of P62 suppressed R848-induced cell death and LC3II protein accumulation. As an important mediator for cell-cell communication, Notch signaling is responsible for cell-fate decisions. Our results showed that activation of TLR7 also upregulated the expression of Notch1, especially its downstream target gene Hairy and enhancer of split 1 (Hes-1) in macrophages. Of note, we found that Hes-1, as a transcriptional factor, controlled TLR7-induced autophagy by regulating P62 expression. Furthermore, to confirm the above results in vivo, TLR7 agonist imiquimod (IMQ)-induced lupus mouse model was prepared. Splenic macrophages from IMQ-treated mice exhibited increased autophagy and cell death as well as enhanced expressions of Notch1 and Hes-1. Our results indicate that Notch1-Hes-1 signaling controls TLR7-induced autophagic death of macrophage via regulation of P62 in mice with lupus.
Figures


Similar articles
-
CD180 Ligation Inhibits TLR7- and TLR9-Mediated Activation of Macrophages and Dendritic Cells Through the Lyn-SHP-1/2 Axis in Murine Lupus.Front Immunol. 2018 Nov 15;9:2643. doi: 10.3389/fimmu.2018.02643. eCollection 2018. Front Immunol. 2018. PMID: 30498494 Free PMC article.
-
Urokinase-type plasminogen activator receptor is required for impairing toll-like receptor 7 signaling on macrophage efferocytosis in lupus.Mol Immunol. 2020 Nov;127:38-45. doi: 10.1016/j.molimm.2020.08.018. Epub 2020 Sep 7. Mol Immunol. 2020. PMID: 32911323
-
B cell autophagy mediates TLR7-dependent autoimmunity and inflammation.Autophagy. 2015;11(7):1010-24. doi: 10.1080/15548627.2015.1052206. Autophagy. 2015. PMID: 26120731 Free PMC article.
-
MDSC-derived S100A8/9 contributes to lupus pathogenesis by promoting TLR7-mediated activation of macrophages and dendritic cells.Cell Mol Life Sci. 2024 Mar 1;81(1):110. doi: 10.1007/s00018-024-05155-w. Cell Mol Life Sci. 2024. PMID: 38429401 Free PMC article.
-
Toll-like receptor 7 agonist imiquimod prevents the progression of SLE in MRL/lpr mice via inhibiting the differentiation of T follicular helper cells.Int Immunopharmacol. 2020 Mar;80:106239. doi: 10.1016/j.intimp.2020.106239. Epub 2020 Jan 30. Int Immunopharmacol. 2020. PMID: 32007709
Cited by
-
Acetylation-Dependent Regulation of Notch Signaling in Macrophages by SIRT1 Affects Sepsis Development.Front Immunol. 2018 May 7;9:762. doi: 10.3389/fimmu.2018.00762. eCollection 2018. Front Immunol. 2018. PMID: 29867921 Free PMC article.
-
Assessing Autophagy in Mouse Models and Patients with Systemic Autoimmune Diseases.Cells. 2017 Jun 28;6(3):16. doi: 10.3390/cells6030016. Cells. 2017. PMID: 28657591 Free PMC article. Review.
-
Interactions of Autophagy and the Immune System in Health and Diseases.Autophagy Rep. 2022;1(1):438-515. doi: 10.1080/27694127.2022.2119743. Epub 2022 Oct 5. Autophagy Rep. 2022. PMID: 37425656 Free PMC article.
-
Strategies of Targeting Inflammasome in the Treatment of Systemic Lupus Erythematosus.Front Immunol. 2022 May 18;13:894847. doi: 10.3389/fimmu.2022.894847. eCollection 2022. Front Immunol. 2022. PMID: 35664004 Free PMC article. Review.
-
Podocytes and autophagy: a potential therapeutic target in lupus nephritis.Autophagy. 2019 May;15(5):908-912. doi: 10.1080/15548627.2019.1580512. Epub 2019 Feb 17. Autophagy. 2019. PMID: 30755075 Free PMC article.
References
-
- Yu C, Gershwin ME, Chang C. Diagnostic criteria for systemic lupus erythematosus: a critical review. J Autoimmun 2014; 48-49: 10–13. - PubMed
-
- Orme J, Mohan C. Macrophage subpopulations in systemic lupus erythematosus. Discov Med 2012; 13: 151–158. - PubMed
-
- Chalmers SA, Chitu V, Ramanujam M, Putterman C. Therapeutic targeting of macrophages in lupus nephritis. Discov Med 2015; 20: 43–49. - PubMed
-
- Burbano C, Vasquez G, Rojas M. Modulatory effects of CD14+CD16++ monocytes on CD14++CD16- monocytes: a possible explanation of monocyte alterations in systemic lupus erythematosus. Arthritis Rheumatol 2014; 66: 3371–3381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

