Insights into newly discovered marks and readers of epigenetic information
- PMID: 27538025
- PMCID: PMC5116920
- DOI: 10.1038/nchembio.2149
Insights into newly discovered marks and readers of epigenetic information
Abstract
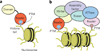
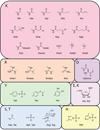
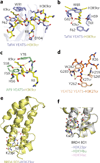
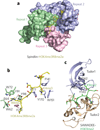
The field of chromatin biology has been advancing at an accelerated pace. Recent discoveries of previously uncharacterized sites and types of post-translational modifications (PTMs) and the identification of new sets of proteins responsible for the deposition, removal, and reading of these marks continue raising the complexity of an already exceedingly complicated biological phenomenon. In this Perspective article we examine the biological importance of new types and sites of histone PTMs and summarize the molecular mechanisms of chromatin engagement by newly discovered epigenetic readers. We also highlight the imperative role of structural insights in understanding PTM-reader interactions and discuss future directions to enhance the knowledge of PTM readout.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

