Structure-Function Analysis of the Two-Peptide Bacteriocin Plantaricin EF
- PMID: 27538436
- PMCID: PMC5026404
- DOI: 10.1021/acs.biochem.6b00588
Structure-Function Analysis of the Two-Peptide Bacteriocin Plantaricin EF
Abstract
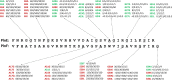
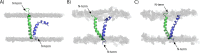
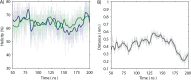
Plantaricin EF is a two-peptide bacteriocin that depends on the complementary action of two different peptides (PlnE and PlnF) to function. The structures of the individual peptides have previously been analyzed by nuclear magnetic resonance spectroscopy ( Fimland, N. et al. ( 2008 ) , Biochim. Biophys. Acta 1784 , 1711 - 1719 ), but the bacteriocin structure and how the two peptides interact have not been determined. All two-peptide bacteriocins identified so far contain GxxxG motifs. These motifs, together with GxxxG-like motifs, are known to mediate helix-helix interactions in membrane proteins. We have mutated all GxxxG and GxxxG-like motifs in PlnE and PlnF in order to determine if any of these motifs are important for antimicrobial activity and thus possibly for interactions between PlnE and PlnF. Moreover, the aromatic amino acids Tyr and Trp in PlnE and PlnF were substituted, and four fusion polypeptides were constructed in order to investigate the relative orientation of PlnE and PlnF in target cell membranes. The results obtained with the fusion polypeptides indicate that PlnE and PlnF interact in an antiparallel manner and that the C-terminus of PlnE and N-terminus of PlnF are on the outer part of target cell membranes and the N-terminus of PlnE and C-terminus of PlnF are on the inner part. The preference for an aromatic residue at position 6 in PlnE suggests a positioning of this residue in or near the membrane interface on the cells inside. Mutations in the GxxxG motifs indicate that the G5xxxG9 motif in PlnE and the S26xxxG30 motif in PlnF are involved in helix-helix interactions. Atomistic molecular dynamics simulation of a structural model consistent with the results confirmed the stability of the structure and its orientation in membranes. The simulation approved the anticipated interactions and revealed additional interactions that further increase the stability of the proposed structure.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF.Biochim Biophys Acta. 2008 Nov;1784(11):1711-9. doi: 10.1016/j.bbapap.2008.05.003. Epub 2008 May 24. Biochim Biophys Acta. 2008. PMID: 18555030
-
Interactions of a class IIb bacteriocin with a model lipid bilayer, investigated through molecular dynamics simulations.Biochim Biophys Acta. 2016 Apr;1858(4):824-35. doi: 10.1016/j.bbamem.2016.01.005. Epub 2016 Jan 13. Biochim Biophys Acta. 2016. PMID: 26774214
-
Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K.J Bacteriol. 1999 Feb;181(3):740-7. doi: 10.1128/JB.181.3.740-747.1999. J Bacteriol. 1999. PMID: 9922235 Free PMC article.
-
The two-peptide class II bacteriocins: structure, production, and mode of action.J Mol Microbiol Biotechnol. 2007;13(4):210-9. doi: 10.1159/000104750. J Mol Microbiol Biotechnol. 2007. PMID: 17827971 Review.
-
An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum.Peptides. 2009 Aug;30(8):1562-74. doi: 10.1016/j.peptides.2009.05.014. Epub 2009 May 22. Peptides. 2009. PMID: 19465075 Review.
Cited by
-
Overview of Global Trends in Classification, Methods of Preparation and Application of Bacteriocins.Antibiotics (Basel). 2020 Aug 28;9(9):553. doi: 10.3390/antibiotics9090553. Antibiotics (Basel). 2020. PMID: 32872235 Free PMC article. Review.
-
Novel Selective Inhibition of Lactobacillus iners by Lactobacillus-Derived Bacteriocins.Appl Environ Microbiol. 2020 Oct 1;86(20):e01594-20. doi: 10.1128/AEM.01594-20. Print 2020 Oct 1. Appl Environ Microbiol. 2020. PMID: 32801180 Free PMC article.
-
Different Combinations of Nitrogen and Carbon Sources Influence the Growth and Postbiotic Metabolite Characteristics of Lactiplantibacillus plantarum Strains Isolated from Malaysian Foods.Foods. 2024 Sep 30;13(19):3123. doi: 10.3390/foods13193123. Foods. 2024. PMID: 39410157 Free PMC article.
-
A novel of new class II bacteriocin from Bacillus velezensis HN-Q-8 and its antibacterial activity on Streptomyces scabies.Front Microbiol. 2022 Jul 29;13:943232. doi: 10.3389/fmicb.2022.943232. eCollection 2022. Front Microbiol. 2022. PMID: 35966655 Free PMC article.
-
Maltose effective improving production and regulatory biosynthesis of plantaricin EF in Lactobacillus plantarum 163.Appl Microbiol Biotechnol. 2021 Apr;105(7):2713-2723. doi: 10.1007/s00253-021-11218-w. Epub 2021 Mar 12. Appl Microbiol Biotechnol. 2021. PMID: 33710357
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

