Transcriptome response of cassava leaves under natural shade
- PMID: 27539510
- PMCID: PMC4990974
- DOI: 10.1038/srep31673
Transcriptome response of cassava leaves under natural shade
Abstract
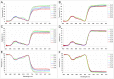
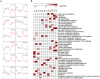
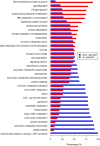
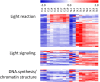
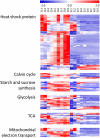
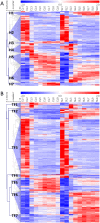
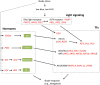
Cassava is an important staple crop in tropical and sub-tropical areas. As a common farming practice, cassava is usually cultivated intercropping with other crops and subjected to various degrees of shading, which causes reduced productivity. Herein, a comparative transcriptomic analysis was performed on a series of developmental cassava leaves under both full sunlight and natural shade conditions. Gene expression profiles of these two conditions exhibited similar developmental transitions, e.g. genes related to cell wall and basic cellular metabolism were highly expressed in immature leaves, genes involved in lipid metabolism and tetrapyrrole synthesis were highly expressed during the transition stages, and genes related to photosynthesis and carbohydrates metabolism were highly expressed in mature leaves. Compared with the control, shade significantly induced the expression of genes involved in light reaction of photosynthesis, light signaling and DNA synthesis/chromatin structure; however, the genes related to anthocyanins biosynthesis, heat shock, calvin cycle, glycolysis, TCA cycle, mitochondrial electron transport, and starch and sucrose metabolisms were dramatically depressed. Moreover, the shade also influenced the expression of hormone-related genes and transcriptional factors. The findings would improve our understanding of molecular mechanisms of shade response, and shed light on pathways associated with shade-avoidance syndrome for cassava improvement.
Figures


Similar articles
-
The Discrepant and Similar Responses of Genome-Wide Transcriptional Profiles between Drought and Cold Stresses in Cassava.Int J Mol Sci. 2017 Dec 12;18(12):2668. doi: 10.3390/ijms18122668. Int J Mol Sci. 2017. PMID: 29231846 Free PMC article.
-
Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress.BMC Genomics. 2012 Feb 10;13:64. doi: 10.1186/1471-2164-13-64. BMC Genomics. 2012. PMID: 22321773 Free PMC article.
-
Transcriptomic profiling suggests candidate molecular responses to waterlogging in cassava.PLoS One. 2022 Jan 21;17(1):e0261086. doi: 10.1371/journal.pone.0261086. eCollection 2022. PLoS One. 2022. PMID: 35061680 Free PMC article.
-
Strand-specific RNA-seq based identification and functional prediction of drought-responsive lncRNAs in cassava.BMC Genomics. 2019 Mar 13;20(1):214. doi: 10.1186/s12864-019-5585-5. BMC Genomics. 2019. PMID: 30866814 Free PMC article.
-
Current knowledge and future research perspectives on cassava (Manihot esculenta Crantz) chemical defenses: An agroecological view.Phytochemistry. 2016 Oct;130:10-21. doi: 10.1016/j.phytochem.2016.05.013. Epub 2016 Jun 14. Phytochemistry. 2016. PMID: 27316676 Review.
Cited by
-
Extensive post-transcriptional regulation revealed by integrative transcriptome and proteome analyses in salicylic acid-induced flowering in duckweed (Lemna gibba).Front Plant Sci. 2024 Feb 7;15:1331949. doi: 10.3389/fpls.2024.1331949. eCollection 2024. Front Plant Sci. 2024. PMID: 38390296 Free PMC article.
-
Physiological and Transcriptomic Analysis Reveals Distorted Ion Homeostasis and Responses in the Freshwater Plant Spirodela polyrhiza L. under Salt Stress.Genes (Basel). 2019 Sep 24;10(10):743. doi: 10.3390/genes10100743. Genes (Basel). 2019. PMID: 31554307 Free PMC article.
-
Monoseeding Increases Peanut (Arachis hypogaea L.) Yield by Regulating Shade-Avoidance Responses and Population Density.Plants (Basel). 2021 Nov 8;10(11):2405. doi: 10.3390/plants10112405. Plants (Basel). 2021. PMID: 34834768 Free PMC article.
-
Co-expression network analysis and cis-regulatory element enrichment determine putative functions and regulatory mechanisms of grapevine ATL E3 ubiquitin ligases.Sci Rep. 2018 Feb 16;8(1):3151. doi: 10.1038/s41598-018-21377-y. Sci Rep. 2018. PMID: 29453355 Free PMC article.
-
Integrated Transcriptome and Metabolic Analyses Reveals Novel Insights into Free Amino Acid Metabolism in Huangjinya Tea Cultivar.Front Plant Sci. 2017 Mar 6;8:291. doi: 10.3389/fpls.2017.00291. eCollection 2017. Front Plant Sci. 2017. PMID: 28321230 Free PMC article.
References
-
- Taylor N., Chavarriaga P., Raemakers K., Siritunga D. & Zhang P. Development and application of transgenic technologies in cassava. Plant Mol. Biol. 56, 671–688 (2004). - PubMed
-
- Souza C. R. B. d., Carvalho L. J. C. B. & Cascardo J. C. d. M. Comparative gene expression study to identify genes possibly related to storage root formation in cassava. Protein Pept. Lett. 11, 577–582 (2004). - PubMed
-
- Koch B. et al.. Possible use of a biotechnological approach to optimize and regulate the content and distribution of cyanogenic glucosides in cassava to increase food safety. In: International Workshop on Cassava Safety 375 (ed^(eds) (1994).
-
- Jansson C., Westerbergh A., Zhang J., Hu X. & Sun C. Cassava, a potential biofuel crop in (the) People’s Republic of China. Applied Energy 86, S95–S99 (2009).
-
- Mutsaers H., Ezumah H. & Osiru D. Cassava-based intercropping: a review. Field Crops Res. 34, 431–457 (1993).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

