Structure-function relationships of archaeal Cbf5 during in vivo RNA-guided pseudouridylation
- PMID: 27539785
- PMCID: PMC5029457
- DOI: 10.1261/rna.057547.116
Structure-function relationships of archaeal Cbf5 during in vivo RNA-guided pseudouridylation
Abstract
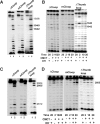
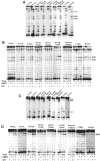
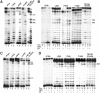
In Eukarya and Archaea, in addition to protein-only pseudouridine (Ψ) synthases, complexes containing one guide RNA and four proteins can also produce Ψ. Cbf5 protein is the Ψ synthase in the complex. Previously, we showed that Ψ's at positions 1940, 1942, and 2605 of Haloferax volcanii 23S rRNA are absent in a cbf5-deleted strain, and a plasmid-borne copy of cbf5 can rescue the synthesis of these Ψ's. Based on published reports of the structure of archaeal Cbf5 complexed with other proteins and RNAs, we identified several potential residues and structures in H. volcanii Cbf5, which were expected to play important roles in pseudouridylation. We mutated these structures and determined their effects on Ψ production at the three rRNA positions under in vivo conditions. Mutations of several residues in the catalytic domain and certain residues in the thumb loop either abolished Ψ's or produced partial modification; the latter indicates a slower rate of Ψ formation. The universal catalytic aspartate of Ψ synthases could be replaced by glutamate in Cbf5. A conserved histidine, which is common to Cbf5 and TruB is not needed, but another conserved histidine of Cbf5 is required for the in vivo RNA-guided Ψ formation. We also identified a previously unreported novelty in the pseudouridylation activity of Cbf5 where a single stem-loop of a guide H/ACA RNA is used to produce two closely placed Ψ's and mutations of certain residues of Cbf5 abolished one of these two Ψ's. In summary, this first in vivo study identifies several structures of an archaeal Cbf5 protein that are important for its RNA-guided pseudouridylation activity.
Keywords: H/ACA RNA; Haloferax volcanii; RNA modification; mutagenesis; protein structure; ribonucleoprotein.
© 2016 Majumder et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
Similar articles
-
Contribution of protein Gar1 to the RNA-guided and RNA-independent rRNA:Ψ-synthase activities of the archaeal Cbf5 protein.Sci Rep. 2018 Sep 14;8(1):13815. doi: 10.1038/s41598-018-32164-0. Sci Rep. 2018. PMID: 30218085 Free PMC article.
-
The presence of the ACA box in archaeal H/ACA guide RNAs promotes atypical pseudouridylation.RNA. 2020 Apr;26(4):396-418. doi: 10.1261/rna.073734.119. Epub 2020 Jan 9. RNA. 2020. PMID: 31919243 Free PMC article.
-
Pseudouridine formation in archaeal RNAs: The case of Haloferax volcanii.RNA. 2011 Jul;17(7):1367-80. doi: 10.1261/rna.2712811. Epub 2011 May 31. RNA. 2011. PMID: 21628430 Free PMC article.
-
Posttranscriptional RNA Pseudouridylation.Enzymes. 2017;41:151-167. doi: 10.1016/bs.enz.2017.02.001. Epub 2017 Mar 11. Enzymes. 2017. PMID: 28601221 Free PMC article. Review.
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
Cited by
-
Contribution of protein Gar1 to the RNA-guided and RNA-independent rRNA:Ψ-synthase activities of the archaeal Cbf5 protein.Sci Rep. 2018 Sep 14;8(1):13815. doi: 10.1038/s41598-018-32164-0. Sci Rep. 2018. PMID: 30218085 Free PMC article.
-
The presence of the ACA box in archaeal H/ACA guide RNAs promotes atypical pseudouridylation.RNA. 2020 Apr;26(4):396-418. doi: 10.1261/rna.073734.119. Epub 2020 Jan 9. RNA. 2020. PMID: 31919243 Free PMC article.
-
The human ortholog of archaeal Pus10 produces pseudouridine 54 in select tRNAs where its recognition sequence contains a modified residue.RNA. 2019 Mar;25(3):336-351. doi: 10.1261/rna.068114.118. Epub 2018 Dec 7. RNA. 2019. PMID: 30530625 Free PMC article.
References
-
- Bakin A, Ofengand J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
