Comparison of Newly Assembled Full Length HIV-1 Integrase With Prototype Foamy Virus Integrase: Structure-Function Prospective
- PMID: 27540450
- PMCID: PMC4976072
- DOI: 10.5812/jjm.29773
Comparison of Newly Assembled Full Length HIV-1 Integrase With Prototype Foamy Virus Integrase: Structure-Function Prospective
Abstract
Background: Drug design against human immunodeficiency virus type 1 (HIV-1) integrase through its mechanistic study is of great interest in the area in biological research. The main obstacle in this area is the absence of the full-length crystal structure for HIV-1 integrase to be used as a model. A complete structure, similar to HIV-1 of a prototype foamy virus integrase in complex with DNA, including all conservative residues, is available and has been extensively used in recent investigations.
Objectives: The aim of this study was to determine whether the above model is precisely representative of HIV-1 integrase. This would critically determine the success of any designed drug using the model in deactivation of integrase and AIDS treatment.
Materials and methods: Primarily, a new structure for HIV-1 was constructed, using a crystal structure of prototype foamy virus as the starting structure. The constructed structure of HIV-1 integrase was simultaneously simulated with a prototype foamy virus integrase on a separate occasion.
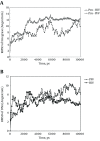
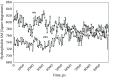
Results: Our results indicate that the HIV-1 system behaves differently from the prototype foamy virus in terms of folding, hydration, hydrophobicity of binding site and stability.
Conclusions: Based on our findings, we can conclude that HIV-1 integrase is vastly different from the prototype foamy virus integrase and does not resemble it, and the modeling output of the prototype foamy virus simulations could not be simply generalized to HIV-1 integrase. Therefore, our HIV-1 model seems to be more representative and more useful for future research.
Keywords: Human Immunodeficiency Virus Type-1; Integrase; Molecular Dynamic Simulation; Prototype Foamy Virus.
Figures



Similar articles
-
Molecular dynamics simulation studies of the wild type and E92Q/N155H mutant of Elvitegravir-resistance HIV-1 integrase.Interdiscip Sci. 2014 Nov 6. doi: 10.1007/s12539-013-0215-4. Online ahead of print. Interdiscip Sci. 2014. PMID: 25373632
-
Study on the interactions between diketo-acid inhibitors and prototype foamy virus integrase-DNA complex via molecular docking and comparative molecular dynamics simulation methods.J Biomol Struct Dyn. 2013;31(7):734-47. doi: 10.1080/07391102.2012.709458. Epub 2012 Aug 22. J Biomol Struct Dyn. 2013. PMID: 22913375
-
Modeling the HIV-1 Intasome: A Prototype View of the Target of Integrase Inhibitors.Viruses. 2010 Dec;2(12):2777-81. doi: 10.3390/v2122777. Epub 2010 Dec 21. Viruses. 2010. PMID: 21994639 Free PMC article.
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
Structure-activity relationships of HIV-1 integrase inhibitors--enzyme-ligand interactions.Curr Med Chem. 2003 Sep;10(18):1795-810. doi: 10.2174/0929867033456981. Curr Med Chem. 2003. PMID: 12871105 Review.
Cited by
-
Chelation Motifs Affecting Metal-dependent Viral Enzymes: N'-acylhydrazone Ligands as Dual Target Inhibitors of HIV-1 Integrase and Reverse Transcriptase Ribonuclease H Domain.Front Microbiol. 2017 Mar 20;8:440. doi: 10.3389/fmicb.2017.00440. eCollection 2017. Front Microbiol. 2017. PMID: 28373864 Free PMC article.
-
Structural Insights on Retroviral DNA Integration: Learning from Foamy Viruses.Viruses. 2019 Aug 22;11(9):770. doi: 10.3390/v11090770. Viruses. 2019. PMID: 31443391 Free PMC article. Review.
References
-
- Feller L, Khammissa RAG., Wood NH, Meyerov R, Lemmer J. Insights into immunopathogenic mechanisms of HIV infection: high levels of immune activation and HIV fitness: communication corner. South African Dental Journal. 2008;63(10):552–7. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
