Molecular Principles of Gene Fusion Mediated Rewiring of Protein Interaction Networks in Cancer
- PMID: 27540857
- PMCID: PMC5003813
- DOI: 10.1016/j.molcel.2016.07.008
Molecular Principles of Gene Fusion Mediated Rewiring of Protein Interaction Networks in Cancer
Abstract
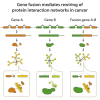
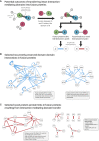
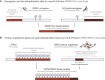
Gene fusions are common cancer-causing mutations, but the molecular principles by which fusion protein products affect interaction networks and cause disease are not well understood. Here, we perform an integrative analysis of the structural, interactomic, and regulatory properties of thousands of putative fusion proteins. We demonstrate that genes that form fusions (i.e., parent genes) tend to be highly connected hub genes, whose protein products are enriched in structured and disordered interaction-mediating features. Fusion often results in the loss of these parental features and the depletion of regulatory sites such as post-translational modifications. Fusion products disproportionately connect proteins that did not previously interact in the protein interaction network. In this manner, fusion products can escape cellular regulation and constitutively rewire protein interaction networks. We suggest that the deregulation of central, interaction-prone proteins may represent a widespread mechanism by which fusion proteins alter the topology of cellular signaling pathways and promote cancer.
Keywords: cancer genomics; fusion protein; gene fusion; protein interaction networks.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Frequent mutations in acetylation and ubiquitination sites suggest novel driver mechanisms of cancer.Genome Med. 2016 May 12;8(1):55. doi: 10.1186/s13073-016-0311-2. Genome Med. 2016. PMID: 27175787 Free PMC article.
-
Fusion proteins mediate alternation of protein interaction networks in cancers.Adv Protein Chem Struct Biol. 2022;131:165-176. doi: 10.1016/bs.apcsb.2022.05.007. Epub 2022 Jun 7. Adv Protein Chem Struct Biol. 2022. PMID: 35871889
-
Do cancer proteins really interact strongly in the human protein-protein interaction network?Comput Biol Chem. 2011 Jun;35(3):121-5. doi: 10.1016/j.compbiolchem.2011.04.005. Comput Biol Chem. 2011. PMID: 21666777 Free PMC article.
-
DAXX in cancer: phenomena, processes, mechanisms and regulation.Nucleic Acids Res. 2019 Sep 5;47(15):7734-7752. doi: 10.1093/nar/gkz634. Nucleic Acids Res. 2019. PMID: 31350900 Free PMC article. Review.
-
Protein interaction networks in medicine and disease.Proteomics. 2012 May;12(10):1706-16. doi: 10.1002/pmic.201100594. Proteomics. 2012. PMID: 22593007 Review.
Cited by
-
FusionPathway: Prediction of pathways and therapeutic targets associated with gene fusions in cancer.PLoS Comput Biol. 2018 Jul 24;14(7):e1006266. doi: 10.1371/journal.pcbi.1006266. eCollection 2018 Jul. PLoS Comput Biol. 2018. PMID: 30040819 Free PMC article.
-
Decoding Oncofusions: Unveiling Mechanisms, Clinical Impact, and Prospects for Personalized Cancer Therapies.Cancers (Basel). 2023 Jul 19;15(14):3678. doi: 10.3390/cancers15143678. Cancers (Basel). 2023. PMID: 37509339 Free PMC article. Review.
-
Construction and analysis of sample-specific driver modules for breast cancer.BMC Genomics. 2022 Oct 20;23(1):717. doi: 10.1186/s12864-022-08928-4. BMC Genomics. 2022. PMID: 36266635 Free PMC article.
-
Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains.Cell. 2018 Dec 13;175(7):1842-1855.e16. doi: 10.1016/j.cell.2018.10.042. Epub 2018 Nov 15. Cell. 2018. PMID: 30449618 Free PMC article.
-
Investigating the disordered regions (MoRFs, SLiMs and LCRs) and functions of mimicry proteins/peptides in silico.PLoS One. 2022 Apr 14;17(4):e0265657. doi: 10.1371/journal.pone.0265657. eCollection 2022. PLoS One. 2022. PMID: 35421114 Free PMC article.
References
-
- Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

