Trans-inactivation: Repression in a wrong place
- PMID: 27540893
- PMCID: PMC5406161
- DOI: 10.1080/19336934.2016.1225634
Trans-inactivation: Repression in a wrong place
Abstract
Trans-inactivation is the repression of genes on a normal chromosome under the influence of a rearranged homologous chromosome demonstrating the position effect variegation (PEV). This phenomenon was studied in detail on the example of brownDominant allele causing the repression of wild-type brown gene on the opposite chromosome. We have investigated another trans-inactivation-inducing chromosome rearrangement, In(2)A4 inversion. In both cases, brownDominant and In(2)A4, the repression seems to be the result of dragging of the euchromatic region of the normal chromosome into the heterochromatic environment. It was found that cis-inactivation (classical PEV) and trans-inactivation show different patterns of distribution along the chromosome and respond differently to PEV modifying genes. It appears that the causative mechanism of trans-inactivation is de novo heterochromatin assembly on euchromatic sequences dragged into the heterochromatic nuclear compartment. Trans-inactivation turns out to be the result of a combination of heterochromatin-induced position effect and the somatic interphase chromosome pairing that is widespread in Diptera.
Keywords: PEV; chromatin; heterochromatin; nuclear compartments; trans-inactivation; transcription.
Figures

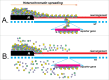
Comment on
- Extra view to: Abramov YA, Shatskikh AS, Maksimenko OG, Bonaccorsi S, Gvozdev VA, Lavrov SA. The Differences Between Cis- and Trans-Gene Inactivation Caused by Heterochromatin in Drosophila. Genetics 2016; 202:93-106.
Similar articles
-
The Differences Between Cis- and Trans-Gene Inactivation Caused by Heterochromatin in Drosophila.Genetics. 2016 Jan;202(1):93-106. doi: 10.1534/genetics.115.181693. Epub 2015 Oct 23. Genetics. 2016. PMID: 26500261 Free PMC article.
-
[Correlation on a cellular level of gene transcriptional silencing and heterochromatin compartment dragging in case of PEV-producing eu-heterochromatin rearrangement in Drosophila melanogaster].Mol Biol (Mosk). 2013 Mar-Apr;47(2):286-91. doi: 10.7868/s0026898413020080. Mol Biol (Mosk). 2013. PMID: 23808163 Russian.
-
The impact of genetic background and cell lineage on the level and pattern of gene expression in position effect variegation.Epigenetics Chromatin. 2019 Nov 13;12(1):70. doi: 10.1186/s13072-019-0314-5. Epigenetics Chromatin. 2019. PMID: 31722719 Free PMC article.
-
Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila.Adv Genet. 2008;61:1-43. doi: 10.1016/S0065-2660(07)00001-6. Adv Genet. 2008. PMID: 18282501 Review.
-
Position effect variegation and chromatin proteins.Bioessays. 1992 Sep;14(9):605-12. doi: 10.1002/bies.950140907. Bioessays. 1992. PMID: 1365916 Review.
Cited by
-
Effects of Chromatin Structure Modifiers on the trans-Acting Heterochromatin Position Effect in Drosophila melanogaster.Dokl Biochem Biophys. 2023 Dec;513(Suppl 1):S75-S81. doi: 10.1134/S160767292470073X. Epub 2024 Feb 20. Dokl Biochem Biophys. 2023. PMID: 38379078
-
Evolutionary Dynamics of the Pericentromeric Heterochromatin in Drosophila virilis and Related Species.Genes (Basel). 2021 Jan 27;12(2):175. doi: 10.3390/genes12020175. Genes (Basel). 2021. PMID: 33513919 Free PMC article.
References
-
- Muller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet 1930; 22:299; http://dx.doi.org/10.1007/BF02984195 - DOI
-
- Girton JR, Johansen KM. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv Genet 2008; 61:1-43; PMID:18282501; http://dx.doi.org/10.1016/S0065-2660(07)00001-6 - DOI - PubMed
-
- Spofford JB. Position-effect variegation in Drosophila. In: Ashburner M, Novitski E (eds.), The genetics and biology of Drosophila, vol 1C. Academic Press, New York, pp 955–1018.
-
- Deng H, Cai W, Wang C, Lerach S, Delattre M, Girton J, Johansen J, Johansen KM. JIL-1 and Su(var)3–7 interact genetically and counteract each other's effect on position-effect variegation in Drosophila. Genetics 2010; 185:1183-92; PMID:20457875; http://dx.doi.org/10.1534/genetics.110.117150 - DOI - PMC - PubMed
-
- Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res 2006; 14:377-92; PMID:16821134; http://dx.doi.org/10.1007/s10577-006-1066-1 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
