Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase
- PMID: 27541856
- PMCID: PMC5103349
- DOI: 10.1080/15548627.2016.1217369
Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase
Abstract
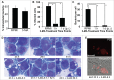
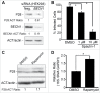
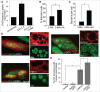
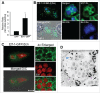
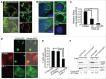
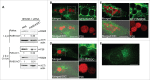
Ehrlichia chaffeensis is an obligatory intracellular bacterium that causes a potentially fatal emerging zoonosis, human monocytic ehrlichiosis. E. chaffeensis has a limited capacity for biosynthesis and metabolism and thus depends mostly on host-synthesized nutrients for growth. Although the host cell cytoplasm is rich with these nutrients, as E. chaffeensis is confined within the early endosome-like membrane-bound compartment, only host nutrients that enter the compartment can be used by this bacterium. How this occurs is unknown. We found that ehrlichial replication depended on autophagy induction involving class III phosphatidylinositol 3-kinase (PtdIns3K) activity, BECN1 (Beclin 1), and ATG5 (autophagy-related 5). Ehrlichia acquired host cell preincorporated amino acids in a class III PtdIns3K-dependent manner and ehrlichial growth was enhanced by treatment with rapamycin, an autophagy inducer. Moreover, ATG5 and RAB5A/B/C were routed to ehrlichial inclusions. RAB5A/B/C siRNA knockdown, or overexpression of a RAB5-specific GTPase-activating protein or dominant-negative RAB5A inhibited ehrlichial infection, indicating the critical role of GTP-bound RAB5 during infection. Both native and ectopically expressed ehrlichial type IV secretion effector protein, Etf-1, bound RAB5 and the autophagy-initiating class III PtdIns3K complex, PIK3C3/VPS34, and BECN1, and homed to ehrlichial inclusions. Ectopically expressed Etf-1 activated class III PtdIns3K as in E. chaffeensis infection and induced autophagosome formation, cleared an aggregation-prone mutant huntingtin protein in a class III PtdIns3K-dependent manner, and enhanced ehrlichial proliferation. These data support the notion that E. chaffeensis secretes Etf-1 to induce autophagy to repurpose the host cytoplasm and capture nutrients for its growth through RAB5 and class III PtdIns3K, while avoiding autolysosomal killing.
Keywords: ATG5; BECN1; Ehrlichia chaffeensis; Etf-1; LC3; RAB5; autophagy; class III PtdIns3K; endosome; infection; type IV secretion effector.
Figures





Similar articles
-
Peptide Nucleic Acid Knockdown and Intra-host Cell Complementation of Ehrlichia Type IV Secretion System Effector.Front Cell Infect Microbiol. 2017 Jun 7;7:228. doi: 10.3389/fcimb.2017.00228. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28638803 Free PMC article.
-
Subversion of RAB5-regulated autophagy by the intracellular pathogen Ehrlichia chaffeensis.Small GTPases. 2019 Sep;10(5):343-349. doi: 10.1080/21541248.2017.1332506. Epub 2017 Jul 5. Small GTPases. 2019. PMID: 28650718 Free PMC article. Review.
-
Ehrlichia type IV secretion system effector Etf-2 binds to active RAB5 and delays endosome maturation.Proc Natl Acad Sci U S A. 2018 Sep 18;115(38):E8977-E8986. doi: 10.1073/pnas.1806904115. Epub 2018 Sep 4. Proc Natl Acad Sci U S A. 2018. PMID: 30181274 Free PMC article.
-
Development of mRNA-lipid nanoparticle intrabodies against rickettsial infection.J Biomed Sci. 2025 Aug 12;32(1):76. doi: 10.1186/s12929-025-01171-5. J Biomed Sci. 2025. PMID: 40797321 Free PMC article.
-
The "Biological Weapons" of Ehrlichia chaffeensis: Novel Molecules and Mechanisms to Subjugate Host Cells.Front Cell Infect Microbiol. 2022 Jan 14;11:830180. doi: 10.3389/fcimb.2021.830180. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35155275 Free PMC article. Review.
Cited by
-
Anaplasma phagocytophilum effector EgeA facilitates infection by hijacking TANGO1 and SCFD1 from ER-Golgi exit sites to pathogen-occupied inclusions.Proc Natl Acad Sci U S A. 2024 Aug 13;121(33):e2405209121. doi: 10.1073/pnas.2405209121. Epub 2024 Aug 6. Proc Natl Acad Sci U S A. 2024. PMID: 39106308 Free PMC article.
-
An intracellular nanobody targeting T4SS effector inhibits Ehrlichia infection.Proc Natl Acad Sci U S A. 2021 May 4;118(18):e2024102118. doi: 10.1073/pnas.2024102118. Proc Natl Acad Sci U S A. 2021. PMID: 33903242 Free PMC article.
-
Ehrlichia Notch signaling induction promotes XIAP stability and inhibits apoptosis.Infect Immun. 2023 Sep 14;91(9):e0000223. doi: 10.1128/iai.00002-23. Epub 2023 Aug 18. Infect Immun. 2023. PMID: 37594275 Free PMC article.
-
Ticks: More Than Just a Pathogen Delivery Service.Front Cell Infect Microbiol. 2021 Sep 1;11:739419. doi: 10.3389/fcimb.2021.739419. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34540723 Free PMC article. No abstract available.
-
Discovery of in vivo Virulence Genes of Obligatory Intracellular Bacteria by Random Mutagenesis.Front Cell Infect Microbiol. 2020 Feb 4;10:2. doi: 10.3389/fcimb.2020.00002. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32117791 Free PMC article.
References
-
- Maeda K, Markowitz N, Hawley RC, Ristic M, Cox D, McDade JE. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med 1987; 316:853–6; PMID:3029590; http://dx.doi.org/10.1056/NEJM198704023161406 - DOI - PubMed
-
- Paddock CD, Childs JE. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev 2003; 16:37–64; PMID:12525424; http://dx.doi.org/10.1128/CMR.16.1.37-64.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
