Computational drugs repositioning identifies inhibitors of oncogenic PI3K/AKT/P70S6K-dependent pathways among FDA-approved compounds
- PMID: 27542212
- PMCID: PMC5312272
- DOI: 10.18632/oncotarget.11318
Computational drugs repositioning identifies inhibitors of oncogenic PI3K/AKT/P70S6K-dependent pathways among FDA-approved compounds
Abstract
The discovery of inhibitors for oncogenic signalling pathways remains a key focus in modern oncology, based on personalized and targeted therapeutics. Computational drug repurposing via the analysis of FDA-approved drug network is becoming a very effective approach to identify therapeutic opportunities in cancer and other human diseases. Given that gene expression signatures can be associated with specific oncogenic mutations, we tested whether a "reverse" oncogene-specific signature might assist in the computational repositioning of inhibitors of oncogenic pathways. As a proof of principle, we focused on oncogenic PI3K-dependent signalling, a molecular pathway frequently driving cancer progression as well as raising resistance to anticancer-targeted therapies. We show that implementation of "reverse" oncogenic PI3K-dependent transcriptional signatures combined with interrogation of drug networks identified inhibitors of PI3K-dependent signalling among FDA-approved compounds. This led to repositioning of Niclosamide (Niclo) and Pyrvinium Pamoate (PP), two anthelmintic drugs, as inhibitors of oncogenic PI3K-dependent signalling. Niclo inhibited phosphorylation of P70S6K, while PP inhibited phosphorylation of AKT and P70S6K, which are downstream targets of PI3K. Anthelmintics inhibited oncogenic PI3K-dependent gene expression and showed a cytostatic effect in vitro and in mouse mammary gland. Lastly, PP inhibited the growth of breast cancer cells harbouring PI3K mutations. Our data indicate that drug repositioning by network analysis of oncogene-specific transcriptional signatures is an efficient strategy for identifying oncogenic pathway inhibitors among FDA-approved compounds. We propose that PP and Niclo should be further investigated as potential therapeutics for the treatment of tumors or diseases carrying the constitutive activation of the PI3K/P70S6K signalling axis.
Keywords: FDA-approved drugs; PI3K-dependent pathways; drugs network; gene expression signatures; oncogenes.
Conflict of interest statement
Authors declare no conflict of interest.
Figures
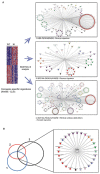
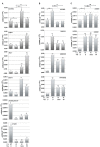

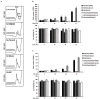
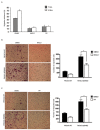
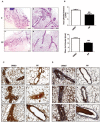
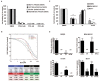
References
-
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. - PubMed
-
- Garraway LA, Janne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. - PubMed
-
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Jr, Marks JR, Dressman HK, West M, Nevins JR. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. - PubMed
-
- Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D'Amico M, Pestell RG, West M, Nevins JR. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. - PubMed
-
- Furge KA, Tan MH, Dykema K, Kort E, Stadler W, Yao X, Zhou M, Teh BT. Identification of deregulated oncogenic pathways in renal cell carcinoma: an integrated oncogenomic approach based on gene expression profiling. Oncogene. 2007;26:1346–1350. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

