Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle
- PMID: 27542410
- PMCID: PMC5076529
- DOI: 10.1074/jbc.M116.749515
Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle
Abstract
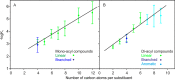
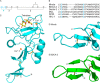
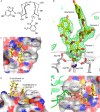
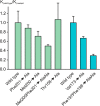
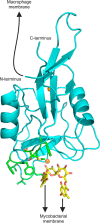
The macrophage receptor mincle binds to trehalose dimycolate on the surface of Mycobacterium tuberculosis Signaling initiated by this interaction leads to cytokine production, which underlies the ability of mycobacteria to evade the immune system and also to function as adjuvants. In previous work the mechanism for binding of the sugar headgroup of trehalose dimycolate to mincle has been elucidated, but the basis for enhanced binding to glycolipid ligands, in which hydrophobic substituents are attached to the 6-hydroxyl groups, has been the subject of speculation. In the work reported here, the interaction of trehalose derivatives with bovine mincle has been probed with a series of synthetic mimics of trehalose dimycolate in binding assays, in structural studies by x-ray crystallography, and by site-directed mutagenesis. Binding studies reveal that, rather than reflecting specific structural preference, the apparent affinity of mincle for ligands with hydrophobic substituents correlates with their overall size. Structural and mutagenesis analysis provides evidence for interaction of the hydrophobic substituents with multiple different portions of the surface of mincle and confirms the presence of three Ca2+-binding sites. The structure of an extended portion of the extracellular domain of mincle, beyond the minimal C-type carbohydrate recognition domain, also constrains the way the binding domains may interact on the surface of macrophages.
Keywords: carbohydrate-binding protein; glycolipid; lectin; mycobacteria; tuberculosis.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

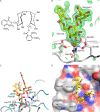
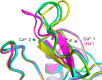
References
-
- Matsumoto M., Tanaka T., Kaisho T., Sanjo H., Copeland N. G., Gilbert D. J., Jenkins N. A., and Akira S. (1999) A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J. Immunol. 163, 5039–5048 - PubMed
-
- Wells C. A., Salvage-Jones J. A., Li X., Hitchens K., Butcher S., Murray R. Z., Beckhouse A. G., Lo Y.-L., Manzanero S., Cobbold C., Schroder K., Ma B., Orr S., Stewart L., Lebus D., et al. (2008) The macrophage-inducible C-type lectin, mincle, is an essential component of the innate immune response to Candida albicans. J. Immunol. 180, 7404–7413 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

