STAMS: STRING-assisted module search for genome wide association studies and application to autism
- PMID: 27542772
- PMCID: PMC5167061
- DOI: 10.1093/bioinformatics/btw530
STAMS: STRING-assisted module search for genome wide association studies and application to autism
Abstract
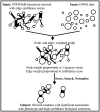
Motivation: Analyzing genome wide association data in the context of biological pathways helps us understand how genetic variation influences phenotype and increases power to find associations. However, the utility of pathway-based analysis tools is hampered by undercuration and reliance on a distribution of signal across all of the genes in a pathway. Methods that combine genome wide association results with genetic networks to infer the key phenotype-modulating subnetworks combat these issues, but have primarily been limited to network definitions with yes/no labels for gene-gene interactions. A recent method (EW_dmGWAS) incorporates a biological network with weighted edge probability by requiring a secondary phenotype-specific expression dataset. In this article, we combine an algorithm for weighted-edge module searching and a probabilistic interaction network in order to develop a method, STAMS, for recovering modules of genes with strong associations to the phenotype and probable biologic coherence. Our method builds on EW_dmGWAS but does not require a secondary expression dataset and performs better in six test cases.
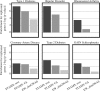
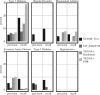
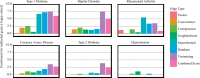
Results: We show that our algorithm improves over EW_dmGWAS and standard gene-based analysis by measuring precision and recall of each method on separately identified associations. In the Wellcome Trust Rheumatoid Arthritis study, STAMS-identified modules were more enriched for separately identified associations than EW_dmGWAS (STAMS P-value 3.0 × 10-4; EW_dmGWAS- P-value = 0.8). We demonstrate that the area under the Precision-Recall curve is 5.9 times higher with STAMS than EW_dmGWAS run on the Wellcome Trust Type 1 Diabetes data.
Availability and implementation: STAMS is implemented as an R package and is freely available at https://simtk.org/projects/stams CONTACT: rbaltman@stanford.eduSupplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures

References
-
- Beissbarth T., Speed T.P. (2004) GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics, 20, 1464–1465. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

