Female mice lacking Xist RNA show partial dosage compensation and survive to term
- PMID: 27542829
- PMCID: PMC5002979
- DOI: 10.1101/gad.281162.116
Female mice lacking Xist RNA show partial dosage compensation and survive to term
Abstract
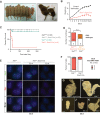
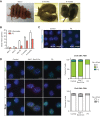
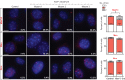
X-chromosome inactivation (XCI) compensates for differences in X-chromosome number between male and female mammals. XCI is orchestrated by Xist RNA, whose expression in early development leads to transcriptional silencing of one X chromosome in the female. Knockout studies have established a requirement for Xist with inviability of female embryos that inherit an Xist deletion from the father. Here, we report that female mice lacking Xist RNA can, surprisingly, develop and survive to term. Xist-null females are born at lower frequency and are smaller at birth, but organogenesis is mostly normal. Transcriptomic analysis indicates significant overexpression of hundreds of X-linked genes across multiple tissues. Therefore, Xist-null mice can develop to term in spite of a deficiency of dosage compensation. However, the degree of X-autosomal dosage imbalance was less than anticipated (1.14-fold to 1.36-fold). Thus, partial dosage compensation can be achieved without Xist, supporting the idea of inherent genome balance. Nevertheless, to date, none of the mutant mice has survived beyond weaning stage. Sudden death is associated with failure of postnatal organ maturation. Our data suggest Xist-independent mechanisms of dosage compensation and demonstrate that small deviations from X-autosomal balance can have profound effects on overall fitness.
Keywords: X inactivation; Xist; dosage compensation; genome balance; inverse effect; knockout mouse; transcriptomics.
© 2016 Yang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




Similar articles
-
The Role of Xist in X-Chromosome Dosage Compensation.Trends Cell Biol. 2018 Dec;28(12):999-1013. doi: 10.1016/j.tcb.2018.05.005. Epub 2018 Jun 14. Trends Cell Biol. 2018. PMID: 29910081 Free PMC article. Review.
-
Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation.Elife. 2020 May 7;9:e54508. doi: 10.7554/eLife.54508. Elife. 2020. PMID: 32379046 Free PMC article.
-
Xist-dependent imprinted X inactivation and the early developmental consequences of its failure.Nat Struct Mol Biol. 2017 Mar;24(3):226-233. doi: 10.1038/nsmb.3365. Epub 2017 Jan 30. Nat Struct Mol Biol. 2017. PMID: 28134930 Free PMC article.
-
Xist RNA in action: Past, present, and future.PLoS Genet. 2019 Sep 19;15(9):e1008333. doi: 10.1371/journal.pgen.1008333. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31537017 Free PMC article. Review.
-
Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation.Development. 2012 Jun;139(12):2130-8. doi: 10.1242/dev.076497. Epub 2012 May 9. Development. 2012. PMID: 22573614 Free PMC article.
Cited by
-
Enhanced chromatin accessibility contributes to X chromosome dosage compensation in mammals.Genome Biol. 2021 Nov 1;22(1):302. doi: 10.1186/s13059-021-02518-5. Genome Biol. 2021. PMID: 34724962 Free PMC article.
-
From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo.Nat Rev Genet. 2022 Apr;23(4):229-243. doi: 10.1038/s41576-021-00427-8. Epub 2021 Nov 26. Nat Rev Genet. 2022. PMID: 34837040 Review.
-
Reverse-genetics studies of lncRNAs-what we have learnt and paths forward.Genome Biol. 2020 Apr 14;21(1):93. doi: 10.1186/s13059-020-01994-5. Genome Biol. 2020. PMID: 32290841 Free PMC article. Review.
-
The Role of Xist in X-Chromosome Dosage Compensation.Trends Cell Biol. 2018 Dec;28(12):999-1013. doi: 10.1016/j.tcb.2018.05.005. Epub 2018 Jun 14. Trends Cell Biol. 2018. PMID: 29910081 Free PMC article. Review.
-
XIST directly regulates X-linked and autosomal genes in naive human pluripotent cells.Cell. 2024 Jan 4;187(1):110-129.e31. doi: 10.1016/j.cell.2023.11.033. Cell. 2024. PMID: 38181737 Free PMC article.
References
-
- Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, Bergmann O. 2015. No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 163: 1026–1036. - PubMed
-
- Birchler JA. 2013. Aneuploidy in plants and flies: the origin of studies of genomic imbalance. Semin Cell Dev Biol 24: 315–319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
