Comparison of Microbial and Chemical Source Tracking Markers To Identify Fecal Contamination Sources in the Humber River (Toronto, Ontario, Canada) and Associated Storm Water Outfalls
- PMID: 27542934
- PMCID: PMC5066352
- DOI: 10.1128/AEM.01675-16
Comparison of Microbial and Chemical Source Tracking Markers To Identify Fecal Contamination Sources in the Humber River (Toronto, Ontario, Canada) and Associated Storm Water Outfalls
Abstract
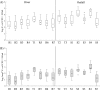
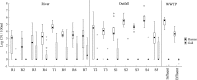
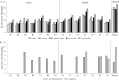
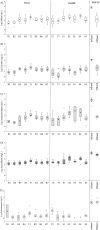
Storm water runoff is a major source of pollution, and understanding the components of storm water discharge is essential to remediation efforts and proper assessment of risks to human and ecosystem health. In this study, culturable Escherichia coli and ampicillin-resistant E. coli levels were quantified and microbial source tracking (MST) markers (including markers for general Bacteroidales spp., human, ruminant/cow, gull, and dog) were detected in storm water outfalls and sites along the Humber River in Toronto, Ontario, Canada, and enumerated via endpoint PCR and quantitative PCR (qPCR). Additionally, chemical source tracking (CST) markers specific for human wastewater (caffeine, carbamazepine, codeine, cotinine, acetaminophen, and acesulfame) were quantified. Human and gull fecal sources were detected at all sites, although concentrations of the human fecal marker were higher, particularly in outfalls (mean outfall concentrations of 4.22 log10 copies, expressed as copy numbers [CN]/100 milliliters for human and 0.46 log10 CN/100 milliliters for gull). Higher concentrations of caffeine, acetaminophen, acesulfame, E. coli, and the human fecal marker were indicative of greater raw sewage contamination at several sites (maximum concentrations of 34,800 ng/liter, 5,120 ng/liter, 9,720 ng/liter, 5.26 log10 CFU/100 ml, and 7.65 log10 CN/100 ml, respectively). These results indicate pervasive sewage contamination at storm water outfalls and throughout the Humber River, with multiple lines of evidence identifying Black Creek and two storm water outfalls with prominent sewage cross-connection problems requiring remediation. Limited data are available on specific sources of pollution in storm water, though our results indicate the value of using both MST and CST methodologies to more reliably assess sewage contamination in impacted watersheds.
Importance: Storm water runoff is one of the most prominent non-point sources of biological and chemical contaminants which can potentially degrade water quality and pose risks to human and ecosystem health. Therefore, identifying fecal contamination in storm water runoff and outfalls is essential for remediation efforts to reduce risks to public health. This study employed multiple methods of identifying levels and sources of fecal contamination in both river and storm water outfall sites, evaluating the efficacy of using culture-based enumeration of E. coli, molecular methods of determining the source(s) of contamination, and CST markers as indicators of fecal contamination. The results identified pervasive human sewage contamination in storm water outfalls and throughout an urban watershed and highlight the utility of using both MST and CST to identify raw sewage contamination.
© Crown copyright 2016.
Figures
Similar articles
-
Microbial Source Tracking Using Quantitative and Digital PCR To Identify Sources of Fecal Contamination in Stormwater, River Water, and Beach Water in a Great Lakes Area of Concern.Appl Environ Microbiol. 2018 Oct 1;84(20):e01634-18. doi: 10.1128/AEM.01634-18. Print 2018 Oct 15. Appl Environ Microbiol. 2018. PMID: 30097445 Free PMC article.
-
Comparative microbial source tracking methods for identification of fecal contamination sources at Sunnyside Beach in the Toronto region area of concern.J Water Health. 2016 Oct;14(5):839-850. doi: 10.2166/wh.2016.296. J Water Health. 2016. PMID: 27740549
-
Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment.Water Res. 2011 Aug;45(14):4081-91. doi: 10.1016/j.watres.2011.04.049. Epub 2011 May 10. Water Res. 2011. PMID: 21689838
-
Sources, pathways, and relative risks of contaminants in surface water and groundwater: a perspective prepared for the Walkerton inquiry.J Toxicol Environ Health A. 2002 Jan 11;65(1):1-142. doi: 10.1080/152873902753338572. J Toxicol Environ Health A. 2002. PMID: 11809004 Review.
-
Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes.FEMS Microbiol Rev. 2014 Jan;38(1):1-40. doi: 10.1111/1574-6976.12031. Epub 2013 Aug 7. FEMS Microbiol Rev. 2014. PMID: 23815638 Review.
Cited by
-
A multi-tiered approach to assess fecal pollution in an urban watershed: Bacterial and viral indicators and sediment microbial communities.Sci Total Environ. 2024 Oct 1;945:174141. doi: 10.1016/j.scitotenv.2024.174141. Epub 2024 Jun 18. Sci Total Environ. 2024. PMID: 38901597
-
Enhanced insights from human and animal host-associated molecular marker genes in a freshwater lake receiving wet weather overflows.Sci Rep. 2019 Aug 29;9(1):12503. doi: 10.1038/s41598-019-48682-4. Sci Rep. 2019. PMID: 31467317 Free PMC article.
-
An overview of molecular markers for identification of non-human fecal pollution sources.Front Microbiol. 2023 Nov 23;14:1256174. doi: 10.3389/fmicb.2023.1256174. eCollection 2023. Front Microbiol. 2023. PMID: 38075863 Free PMC article. Review.
-
Fecal source tracking and eDNA profiling in an urban creek following an extreme rain event.Sci Rep. 2018 Sep 26;8(1):14390. doi: 10.1038/s41598-018-32680-z. Sci Rep. 2018. PMID: 30258068 Free PMC article.
-
Aquatic microbial diversity associated with faecal pollution of Norwegian waterbodies characterized by 16S rRNA gene amplicon deep sequencing.Microb Biotechnol. 2019 Nov;12(6):1487-1491. doi: 10.1111/1751-7915.13461. Epub 2019 Jul 9. Microb Biotechnol. 2019. PMID: 31290258 Free PMC article.
References
-
- Sidhu JPS, Ahmed W, Gernjak W, Aryal R, McCarthy D, Palmer A, Kolotelo P, Toze S. 2013. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci Total Environ 463–464:488–496. - PubMed
-
- Brownell MJ, Harwood VJ, Kurz RC, McQuaig SM, Lukasik J, Scott TM. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res 41:3747–3757. doi:10.1016/j.watres.2007.04.001. - DOI - PubMed
-
- Dorfman M. 2006. Testing the waters: a guide to water quality at vacation beaches. National Resource Defense Council, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

