Role of Phosphate Transport System Component PstB1 in Phosphate Internalization by Nostoc punctiforme
- PMID: 27542935
- PMCID: PMC5066351
- DOI: 10.1128/AEM.01336-16
Role of Phosphate Transport System Component PstB1 in Phosphate Internalization by Nostoc punctiforme
Abstract
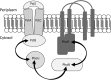
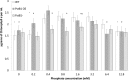
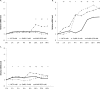
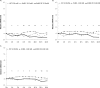
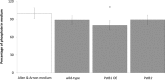
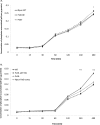
In bacteria, limited phosphate availability promotes the synthesis of active uptake systems, such as the Pst phosphate transport system. To understand the mechanisms that facilitate phosphate accumulation in the cyanobacterium Nostoc punctiforme, phosphate transport systems were identified, revealing a redundancy of Pst phosphate uptake systems that exists across three distinct operons. Four separate PstB system components were identified. pstB1 was determined to be a suitable target for creating phenotypic mutations that could result in the accumulation of excessive levels of phosphate through its overexpression or in a reduction of the capacity to accumulate phosphate through its deletion. Using quantitative real-time PCR (qPCR), it was determined that pstB1 mRNA levels increased significantly over 64 h in cells cultured in 0 mM added phosphate and decreased significantly in cells exposed to high (12.8 mM) phosphate concentrations compared to the level in cells cultured under normal (0.8 mM) conditions. Possible compensation for the loss of PstB1 was observed when pstB2, pstB3, and pstB4 mRNA levels increased, particularly in cells starved of phosphate. The overexpression of pstB1 increased phosphate uptake by N. punctiforme and was shown to functionally complement the loss of PstB in E. coli PstB knockout (PstB-) mutants. The knockout of pstB1 in N. punctiforme did not have a significant effect on cellular phosphate accumulation or growth for the most part, which is attributed to the compensation for the loss of PstB1 by alterations in the pstB2, pstB3, and pstB4 mRNA levels. This study provides novel in vivo evidence that PstB1 plays a functional role in phosphate uptake in N. punctiforme IMPORTANCE: Cyanobacteria have been evolving over 3.5 billion years and have become highly adept at growing under limiting nutrient levels. Phosphate is crucial for the survival and prosperity of all organisms. In bacteria, limited phosphate availability promotes the synthesis of active uptake systems. The Pst phosphate transport system is one such system, responsible for the internalization of phosphate when cells are in phosphate-limited environments. Our investigations reveal the presence of multiple Pst phosphate uptake systems that exist across three distinct operons in Nostoc punctiforme and functionally characterize the role of the gene product PstB1 as being crucial for the maintenance of phosphate accumulation. We demonstrate that the genes pstB2, pstB3, and pstB4 show alterations in expression to compensate for the deletion of pstB1 The overall outcomes of this work provide insights as to the complex transport mechanisms that exist in cyanobacteria like N. punctiforme, allowing them to thrive in low-phosphate environments.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
The adjacent ATP-binding protein-encoding genes of the Enterococcus faecalis phosphate-specific transport (pst) locus have non-overlapping cellular functions.J Bacteriol. 2025 May 22;207(5):e0003325. doi: 10.1128/jb.00033-25. Epub 2025 Apr 14. J Bacteriol. 2025. PMID: 40227050 Free PMC article.
-
Molecular and cellular characterisation of the zinc uptake (Znu) system of Nostoc punctiforme.FEMS Microbiol Ecol. 2013 Nov;86(2):149-71. doi: 10.1111/1574-6941.12153. Epub 2013 Jun 19. FEMS Microbiol Ecol. 2013. PMID: 23710564
-
The ZntA-like NpunR4017 plays a key role in maintaining homeostatic levels of zinc in Nostoc punctiforme.Appl Microbiol Biotechnol. 2015 Dec;99(24):10559-74. doi: 10.1007/s00253-015-6922-5. Epub 2015 Aug 20. Appl Microbiol Biotechnol. 2015. PMID: 26290176
-
Cellular differentiation in the cyanobacterium Nostoc punctiforme.Arch Microbiol. 2002 Dec;178(6):395-403. doi: 10.1007/s00203-002-0476-5. Epub 2002 Sep 18. Arch Microbiol. 2002. PMID: 12420158 Review.
-
The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis.FEMS Microbiol Rev. 2008 May;32(3):461-73. doi: 10.1111/j.1574-6976.2008.00101.x. Epub 2008 Jan 29. FEMS Microbiol Rev. 2008. PMID: 18248418 Review.
Cited by
-
Inhibition Mechanism of Lactiplantibacillus plantarum on the Growth and Biogenic Amine Production in Morganella morganii.Foods. 2023 Sep 29;12(19):3625. doi: 10.3390/foods12193625. Foods. 2023. PMID: 37835277 Free PMC article.
-
Whole genome analysis of endophytic strain PM1 reveals promising plant Growth-Promoting mechanisms in pomegranate.J Genet Eng Biotechnol. 2025 Jun;23(2):100486. doi: 10.1016/j.jgeb.2025.100486. Epub 2025 Apr 5. J Genet Eng Biotechnol. 2025. PMID: 40390499 Free PMC article.
-
Getting Grip on Phosphorus: Potential of Microalgae as a Vehicle for Sustainable Usage of This Macronutrient.Plants (Basel). 2024 Jul 3;13(13):1834. doi: 10.3390/plants13131834. Plants (Basel). 2024. PMID: 38999674 Free PMC article. Review.
-
Consortium of plant growth-promoting rhizobacteria enhances oilseed rape (Brassica napus L.) growth under normal and saline conditions.Arch Microbiol. 2022 Jun 15;204(7):393. doi: 10.1007/s00203-022-03018-1. Arch Microbiol. 2022. PMID: 35704071
-
Role of phosphate sensing in bone and mineral metabolism.Nat Rev Endocrinol. 2018 Nov;14(11):637-655. doi: 10.1038/s41574-018-0076-3. Nat Rev Endocrinol. 2018. PMID: 30218014 Free PMC article. Review.
References
-
- Ann Y, Reddy KR, Delfino JJ. 1999. Influence of chemical amendments on phosphorus immobilization in soils from a constructed wetland. Ecol Eng 14:157–167. doi: 10.1016/S0925-8574(99)00026-9. - DOI
-
- Dokulil M, Teubner K. 2000. Cyanobacterial dominance in lakes. Hydrobiologia 438:1–12. doi: 10.1023/A:1004155810302. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

