UAP56 is a conserved crucial component of a divergent mRNA export pathway in Toxoplasma gondii
- PMID: 27542978
- PMCID: PMC5118106
- DOI: 10.1111/mmi.13485
UAP56 is a conserved crucial component of a divergent mRNA export pathway in Toxoplasma gondii
Abstract
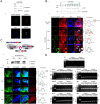
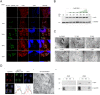
Nucleo-cytoplasmic RNA export is an essential post-transcriptional step to control gene expression in eukaryotic cells and is poorly understood in apicomplexan parasites. With the exception of UAP56, a component of TREX (Transcription Export) complex, other components of mRNA export machinery are not well conserved in divergent supergroups. Here, we use Toxoplasma gondii as a model system to functionally characterize TgUAP56 and its potential interaction factors. We demonstrate that TgUAP56 is crucial for mRNA export and that functional interference leads to significant accumulation of mRNA in the nucleus. It was necessary to employ bioinformatics and phylogenetic analysis to identify orthologs related to mRNA export, which show a remarkable low level of conservation in T. gondii. We adapted a conditional Cas9/CRISPR system to carry out a genetic screen to verify if these factors were involved in mRNA export in T. gondii. Only the disruption of TgRRM_1330 caused accumulation of mRNA in the nucleus as found with TgUAP56. This protein is potentially a divergent partner of TgUAP56, and provides insight into a divergent mRNA export pathway in apicomplexans.
© 2016 The Authors. Molecular Microbiology Published by John Wiley & Sons Ltd.
Figures



Similar articles
-
mRNA export in the apicomplexan parasite Toxoplasma gondii: emerging divergent components of a crucial pathway.Parasit Vectors. 2018 Jan 25;11(1):62. doi: 10.1186/s13071-018-2648-4. Parasit Vectors. 2018. PMID: 29370868 Free PMC article. Review.
-
ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex.Genes Dev. 2010 Sep 15;24(18):2043-53. doi: 10.1101/gad.1898610. Genes Dev. 2010. PMID: 20844015 Free PMC article.
-
The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression.Mol Biol Cell. 2010 Aug 15;21(16):2953-65. doi: 10.1091/mbc.E09-10-0913. Epub 2010 Jun 23. Mol Biol Cell. 2010. PMID: 20573985 Free PMC article.
-
Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication.PLoS Pathog. 2008 Oct;4(10):e1000194. doi: 10.1371/journal.ppat.1000194. Epub 2008 Oct 31. PLoS Pathog. 2008. PMID: 18974867 Free PMC article.
-
Dbp5 - from nuclear export to translation.Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
-
Sending the message: specialized RNA export mechanisms in trypanosomes.Trends Parasitol. 2022 Oct;38(10):854-867. doi: 10.1016/j.pt.2022.07.008. Epub 2022 Aug 24. Trends Parasitol. 2022. PMID: 36028415 Free PMC article. Review.
-
Development of a CRISPR/Cas9 system in Entamoeba histolytica: proof of concept.Int J Parasitol. 2021 Feb;51(2-3):193-200. doi: 10.1016/j.ijpara.2020.09.005. Epub 2020 Nov 29. Int J Parasitol. 2021. PMID: 33264648 Free PMC article.
-
Identification of the Toxoplasma gondii mitochondrial ribosome, and characterisation of a protein essential for mitochondrial translation.Mol Microbiol. 2019 Oct;112(4):1235-1252. doi: 10.1111/mmi.14357. Epub 2019 Jul 24. Mol Microbiol. 2019. PMID: 31339607 Free PMC article.
-
mRNA export in the apicomplexan parasite Toxoplasma gondii: emerging divergent components of a crucial pathway.Parasit Vectors. 2018 Jan 25;11(1):62. doi: 10.1186/s13071-018-2648-4. Parasit Vectors. 2018. PMID: 29370868 Free PMC article. Review.
-
Roles and Cellular Localization of GBP2 and NAB2 During the Blood Stage of Malaria Parasites.Front Cell Infect Microbiol. 2021 Sep 15;11:737457. doi: 10.3389/fcimb.2021.737457. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34604117 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

