Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage
- PMID: 27543075
- PMCID: PMC5137425
- DOI: 10.1093/nar/gkw719
Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage
Abstract
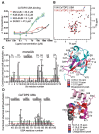
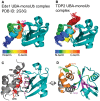
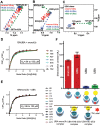
Tyrosyl DNA phosphodiesterase 2 (TDP2) is a multifunctional protein implicated in DNA repair, signal transduction and transcriptional regulation. In its DNA repair role, TDP2 safeguards genome integrity by hydrolyzing 5'-tyrosyl DNA adducts formed by abortive topoisomerase II (Top2) cleavage complexes to allow error-free repair of DNA double-strand breaks, thereby conferring cellular resistance against Top2 poisons. TDP2 consists of a C-terminal catalytic domain responsible for its phosphodiesterase activity, and a functionally uncharacterized N-terminal region. Here, we demonstrate that this N-terminal region contains a ubiquitin (Ub)-associated (UBA) domain capable of binding multiple forms of Ub with distinct modes of interactions and preference for either K48- or K63-linked polyUbs over monoUb. The structure of TDP2 UBA bound to monoUb shows a canonical mode of UBA-Ub interaction. However, the absence of the highly conserved MGF motif and the presence of a fourth α-helix make TDP2 UBA distinct from other known UBAs. Mutations in the TDP2 UBA-Ub binding interface do not affect nuclear import of TDP2, but severely compromise its ability to repair Top2-mediated DNA damage, thus establishing the importance of the TDP2 UBA-Ub interaction in DNA repair. The differential binding to multiple Ub forms could be important for responding to DNA damage signals under different contexts or to support the multi-functionality of TDP2.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.Nucleic Acids Res. 2020 Jun 19;48(11):6310-6325. doi: 10.1093/nar/gkaa318. Nucleic Acids Res. 2020. PMID: 32356875 Free PMC article.
-
Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation.Nucleic Acids Res. 2018 Jan 25;46(2):520-537. doi: 10.1093/nar/gkx1219. Nucleic Acids Res. 2018. PMID: 29216365 Free PMC article. Review.
-
Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2.Nucleic Acids Res. 2016 May 5;44(8):3829-44. doi: 10.1093/nar/gkw228. Epub 2016 Apr 8. Nucleic Acids Res. 2016. PMID: 27060144 Free PMC article.
-
Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing.DNA Repair (Amst). 2016 Jul;43:38-47. doi: 10.1016/j.dnarep.2016.04.009. Epub 2016 May 7. DNA Repair (Amst). 2016. PMID: 27235629
-
Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA.Genes (Basel). 2019 Oct 30;10(11):868. doi: 10.3390/genes10110868. Genes (Basel). 2019. PMID: 31671674 Free PMC article. Review.
Cited by
-
Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.Nucleic Acids Res. 2020 Jun 19;48(11):6310-6325. doi: 10.1093/nar/gkaa318. Nucleic Acids Res. 2020. PMID: 32356875 Free PMC article.
-
Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation.Nucleic Acids Res. 2018 Jan 25;46(2):520-537. doi: 10.1093/nar/gkx1219. Nucleic Acids Res. 2018. PMID: 29216365 Free PMC article. Review.
-
Topoisomerase-Mediated DNA Damage in Neurological Disorders.Front Aging Neurosci. 2021 Nov 25;13:751742. doi: 10.3389/fnagi.2021.751742. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34899270 Free PMC article. Review.
-
Characterization of N- and C-terminal endogenously tagged Tyrosyl-DNA phosphodiesterase 2 (TDPT-1) C. elegans strains.MicroPubl Biol. 2022 Mar 17;2022:10.17912/micropub.biology.000540. doi: 10.17912/micropub.biology.000540. eCollection 2022. MicroPubl Biol. 2022. PMID: 35622466 Free PMC article.
-
Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution.Cell Mol Life Sci. 2020 Jan;77(1):81-91. doi: 10.1007/s00018-019-03367-z. Epub 2019 Nov 15. Cell Mol Life Sci. 2020. PMID: 31728578 Free PMC article. Review.
References
-
- Cortes Ledesma F., El Khamisy S.F., Zuma M.C., Osborn K., Caldecott K.W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. - PubMed
-
- Pei H., Yordy J.S., Leng Q., Zhao Q., Watson D.K., Li R. EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene. 2003;22:2699–2709. - PubMed
-
- Pype S., Declercq W., Ibrahimi A., Michiels C., Van Rietschoten J.G., Dewulf N., de Boer M., Vandenabeele P., Huylebroeck D., Remacle J.E. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. Biol. Chem. 2000;275:18586–18593. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

