Mitochondrial Defects Confer Tolerance against Cellulose Deficiency
- PMID: 27543091
- PMCID: PMC5059812
- DOI: 10.1105/tpc.16.00540
Mitochondrial Defects Confer Tolerance against Cellulose Deficiency
Abstract
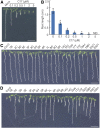
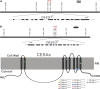
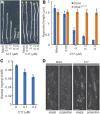
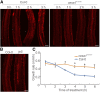
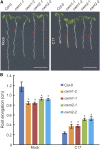
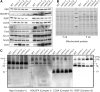
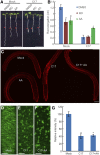
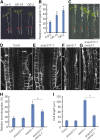
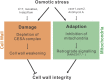
Because the plant cell wall provides the first line of defense against biotic and abiotic assaults, its functional integrity needs to be maintained under stress conditions. Through a phenotype-based compound screening approach, we identified a novel cellulose synthase inhibitor, designated C17. C17 administration depletes cellulose synthase complexes from the plasma membrane in Arabidopsis thaliana, resulting in anisotropic cell elongation and a weak cell wall. Surprisingly, in addition to mutations in CELLULOSE SYNTHASE1 (CESA1) and CESA3, a forward genetic screen identified two independent defective genes encoding pentatricopeptide repeat (PPR)-like proteins (CELL WALL MAINTAINER1 [CWM1] and CWM2) as conferring tolerance to C17. Functional analysis revealed that mutations in these PPR proteins resulted in defective cytochrome c maturation and activation of mitochondrial retrograde signaling, as evidenced by the induction of an alternative oxidase. These mitochondrial perturbations increased tolerance to cell wall damage induced by cellulose deficiency. Likewise, administration of antimycin A, an inhibitor of mitochondrial complex III, resulted in tolerance toward C17. The C17 tolerance of cwm2 was partially lost upon depletion of the mitochondrial retrograde regulator ANAC017, demonstrating that ANAC017 links mitochondrial dysfunction with the cell wall. In view of mitochondria being a major target of a variety of stresses, our data indicate that plant cells might modulate mitochondrial activity to maintain a functional cell wall when subjected to stresses.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. - PubMed
-
- Bartoli C.G., Gómez F., Martínez D.E., Guiamet J.J. (2004). Mitochondria are the main target for oxidative damage in leaves of wheat (Triticum aestivum L.). J. Exp. Bot. 55: 1663–1669. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

