Transcriptome analysis revealed that a quorum sensing system regulates the transfer of the pAt megaplasmid in Agrobacterium tumefaciens
- PMID: 27543103
- PMCID: PMC4992315
- DOI: 10.1186/s12864-016-3007-5
Transcriptome analysis revealed that a quorum sensing system regulates the transfer of the pAt megaplasmid in Agrobacterium tumefaciens
Abstract
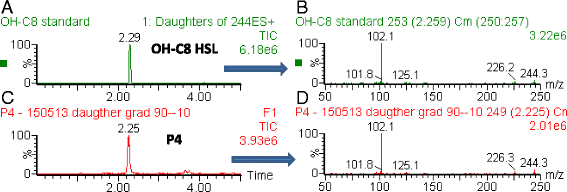
Background: Agrobacterium tumefaciens strain P4 is atypical, as the strain is not pathogenic and produces a for this species unusual quorum sensing signal, identified as N-(3-hydroxy-octanoyl)-homoserine lactone (3OH,C8-HSL).
Results: By sequence analysis and cloning, a functional luxI-like gene, named cinI, has been identified on the At plasmid of A. tumefaciens strain P4. Insertion mutagenesis in the cinI gene and transcriptome analyses permitted the identification of 32 cinI-regulated genes in this strain, most of them encoding proteins responsible for the conjugative transfer of pAtP4. Among these genes were the avhB genes that encode a type 4 secretion system (T4SS) involved in the formation of the conjugation apparatus, the tra genes that encode the DNA transfer and replication (Dtr) machinery and cinI and two luxR orthologs. These last two genes, cinR and cinX, exhibit an unusual organization, with the cinI gene surrounded by the two luxR orthologs. Conjugation experiments confirmed that the conjugative transfer of pAtP4 is regulated by 3OH,C8-HSL. Root colonization experiments indicated that the quorum sensing regulation of the conjugation of the pAtP4 does not confer a gain or a loss of fitness to the bacterial host in the tomato plant rhizosphere.
Conclusion: This work is the first identification of the occurrence of a quorum sensing regulation of the pAt conjugation phenomenon in Agrobacterium.
Keywords: Acylhomoserine lactone; Agrobacterium; At plasmid; Conjugation; Dtr system; Quorum sensing; T4SS; Transcriptomics.
Figures




Similar articles
-
The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci.Mol Microbiol. 2000 Jul;37(1):81-97. doi: 10.1046/j.1365-2958.2000.01960.x. Mol Microbiol. 2000. PMID: 10931307
-
N-acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer.J Bacteriol. 2002 Aug;184(16):4510-9. doi: 10.1128/JB.184.16.4510-4519.2002. J Bacteriol. 2002. PMID: 12142421 Free PMC article.
-
Identification and characterization of a second quorum-sensing system in Agrobacterium tumefaciens A6.J Bacteriol. 2014 Apr;196(7):1403-11. doi: 10.1128/JB.01351-13. Epub 2014 Jan 24. J Bacteriol. 2014. PMID: 24464459 Free PMC article.
-
Cell-cell communication in the plant pathogen Agrobacterium tumefaciens.Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1135-48. doi: 10.1098/rstb.2007.2040. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360279 Free PMC article. Review.
-
Functions and regulation of quorum-sensing in Agrobacterium tumefaciens.Front Plant Sci. 2014 Jan 31;5:14. doi: 10.3389/fpls.2014.00014. eCollection 2014. Front Plant Sci. 2014. PMID: 24550924 Free PMC article. Review.
Cited by
-
Saline Environments as a Source of Potential Quorum Sensing Disruptors to Control Bacterial Infections: A Review.Mar Drugs. 2019 Mar 25;17(3):191. doi: 10.3390/md17030191. Mar Drugs. 2019. PMID: 30934619 Free PMC article. Review.
-
Transcriptome Profiling Reveals the EanI/R Quorum Sensing Regulon in Pantoea Ananatis LMG 2665T.Genes (Basel). 2018 Mar 7;9(3):148. doi: 10.3390/genes9030148. Genes (Basel). 2018. PMID: 29518982 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

