The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment
- PMID: 27543604
- PMCID: PMC5049392
- DOI: 10.1093/jxb/erw307
The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment
Abstract
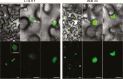
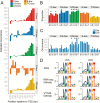
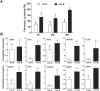
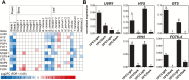
Grapevine (Vitis vinifera L.) is a species well known for its adaptation to radiation. However, photomorphogenic factors related to UV-B responses have not been molecularly characterized. We cloned and studied the role of UV-B RECEPTOR (UVR1), ELONGATED HYPOCOTYL 5 (HY5), and HY5 HOMOLOGUE (HYH) from V. vinifera We performed gene functional characterizations, generated co-expression networks, and tested them in different environmental conditions. These genes complemented the Arabidopsis uvr8 and hy5 mutants in morphological and secondary metabolic responses to radiation. We combined microarray and RNA sequencing (RNA-seq) data with promoter inspections to identify HY5 and HYH putative target genes and their DNA binding preferences. Despite sharing a large set of common co-expressed genes, we found different hierarchies for HY5 and HYH depending on the organ and stress condition, reflecting both co-operative and partially redundant roles. New candidate UV-B gene markers were supported by the presence of HY5-binding sites. These included a set of flavonol-related genes that were up-regulated in a HY5 transient expression assay. We irradiated in vitro plantlets and fruits from old potted vines with high and low UV-B exposures and followed the accumulation of flavonols and changes in gene expression in comparison with non-irradiated conditions. UVR1, HY5, and HYH expression varied with organ, developmental stage, and type of radiation. Surprisingly, UVR1 expression was modulated by shading and temperature in berries, but not by UV-B radiation. We propose that the UV-B response machinery favours berry flavonol accumulation through the activation of HY5 and HYH at different developmental stages at both high and low UV-B exposures.
Keywords: Binding; MYBF1; UVR8.; glycosyltransferase; network; photolyase; ripening.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures





References
-
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. 1998. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Molecular Cell 1, 213–222. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

