Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease
- PMID: 27544059
- PMCID: PMC5226366
- DOI: 10.1016/j.jtho.2016.08.123
Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease
Abstract
Introduction: Low-dose computed tomography (LDCT) is used for screening for lung cancer (LC) in high-risk patients in the United States. The definition of high risk and the impact of frequent false-positive results of low-dose computed tomography remains a challenge. DNA methylation biomarkers are valuable noninvasive diagnostic tools for cancer detection. This study reports on the evaluation of methylation markers in plasma DNA for LC detection and discrimination of malignant from nonmalignant lung disease.
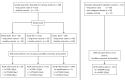
Methods: Circulating DNA was extracted from 3.5-mL plasma samples, treated with bisulfite using a commercially available kit, purified, and assayed by real-time polymerase chain reaction for assessment of DNA methylation of short stature homeobox 2 gene (SHOX2), prostaglandin E receptor 4 gene (PTGER4), and forkhead box L2 gene (FOXL2). In three independent case-control studies these assays were evaluated and optimized. The resultant assay, a triplex polymerase chain reaction combining SHOX2, PTGER4, and the reference gene actin, beta gene (ACTB), was validated using plasma from patients with and without malignant disease.
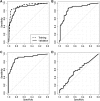
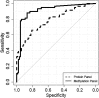
Results: A panel of SHOX2 and PTGER4 provided promising results in three independent case-control studies examining a total of 330 plasma specimens (area under the receiver operating characteristic curve = 91%-98%). A validation study with 172 patient samples demonstrated significant discriminatory performance in distinguishing patients with LC from subjects without malignancy (area under the curve = 0.88). At a fixed specificity of 90%, sensitivity for LC was 67%; at a fixed sensitivity of 90%, specificity was 73%.
Conclusions: Measurement of SHOX2 and PTGER4 methylation in plasma DNA allowed detection of LC and differentiation of nonmalignant diseases. Development of a diagnostic test based on this panel may provide clinical utility in combination with current imaging techniques to improve LC risk stratification.
Keywords: Circulating tumor DNA; DNA methylation; Liquid biopsy; Lung cancer early detection; PTGER4; SHOX2.
Copyright © 2016 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
A Novel Diagnosis Method Based on Methylation Analysis of SHOX2 and Serum Biomarker for Early Stage Lung Cancer.Cancer Control. 2020 Jan-Dec;27(1):1073274820969703. doi: 10.1177/1073274820969703. Cancer Control. 2020. PMID: 33167712 Free PMC article.
-
[DNA methylation detection of SHOX2 and PTGER4 in plasma contributes to differential diagnosis of pulmonary nodule patients].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019 Apr;35(4):357-361. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019. PMID: 31167696 Chinese.
-
SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma.J Thorac Oncol. 2011 Oct;6(10):1632-8. doi: 10.1097/JTO.0b013e318220ef9a. J Thorac Oncol. 2011. PMID: 21694641
-
Association of DNA methylation of RASSF1A and SHOX2 with lung cancer risk: A systematic review and meta-analysis.Medicine (Baltimore). 2024 Dec 13;103(50):e40042. doi: 10.1097/MD.0000000000040042. Medicine (Baltimore). 2024. PMID: 39686414 Free PMC article.
-
Serum Protein Markers for the Early Detection of Lung Cancer: A Focus on Autoantibodies.J Proteome Res. 2017 Jan 6;16(1):3-13. doi: 10.1021/acs.jproteome.6b00559. Epub 2016 Nov 2. J Proteome Res. 2017. PMID: 27769114 Review.
Cited by
-
DNA methylation in human diseases.Heliyon. 2024 Jun 4;10(11):e32366. doi: 10.1016/j.heliyon.2024.e32366. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38933971 Free PMC article. Review.
-
A novel multimodal prediction model based on DNA methylation biomarkers and low-dose computed tomography images for identifying early-stage lung cancer.Chin J Cancer Res. 2023 Oct 30;35(5):511-525. doi: 10.21147/j.issn.1000-9604.2023.05.08. Chin J Cancer Res. 2023. PMID: 37969955 Free PMC article.
-
TMPRSS4: A Novel Tumor Prognostic Indicator for the Stratification of Stage IA Tumors and a Liquid Biopsy Biomarker for NSCLC Patients.J Clin Med. 2019 Dec 3;8(12):2134. doi: 10.3390/jcm8122134. J Clin Med. 2019. PMID: 31817025 Free PMC article.
-
Identification of Hub Genes for Early Diagnosis and Predicting Prognosis in Colon Adenocarcinoma.Biomed Res Int. 2022 Jun 21;2022:1893351. doi: 10.1155/2022/1893351. eCollection 2022. Biomed Res Int. 2022. Retraction in: Biomed Res Int. 2024 Mar 20;2024:9793629. doi: 10.1155/2024/9793629. PMID: 35774271 Free PMC article. Retracted.
-
SHOX CNE9/10 Knockout in U2OS Osteosarcoma Cells and Its Effects on Cell Growth and Apoptosis.Med Sci Monit. 2020 Feb 7;26:e921233. doi: 10.12659/MSM.921233. Med Sci Monit. 2020. PMID: 32032347 Free PMC article.
References
-
- Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. - PubMed
-
- US Centers for Disease Control and Prevention. Lung cancer screening guidelines and recommendations. http://www.cdc.gov/cancer/lung/pdf/guidelines.pdf. Accessed May 5, 2016.
-
- van Klaveren R.J., Oudkerk M., Prokop M. Management of lung nodules detected by volume CT scanning. N Engl J Med. 2009;361:2221–2229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

