Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals
- PMID: 27545045
- PMCID: PMC5025389
- DOI: 10.1016/j.chom.2016.07.015
Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals
Abstract
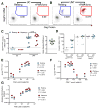
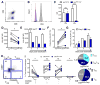
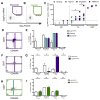
HIV cure efforts are hampered by limited characterization of the cells supporting HIV replication in vivo and inadequate methods for quantifying the latent viral reservoir in patients receiving antiretroviral therapy. We combine fluorescent in situ RNA hybridization with detection of HIV protein and flow cytometry, enabling detection of 0.5-1 gag-pol mRNA(+)/Gag protein(+)-infected cells per million. In the peripheral blood of untreated persons, active HIV replication correlated with viremia and occurred in CD4 T cells expressing T follicular helper cell markers and inhibitory co-receptors. In virally suppressed subjects, the approach identified latently infected cells capable of producing HIV mRNA and protein after stimulation with PMA/ionomycin and latency-reversing agents (LRAs). While ingenol-induced reactivation mirrored the effector and central/transitional memory CD4 T cell contribution to the pool of integrated HIV DNA, bryostatin-induced reactivation occurred predominantly in cells expressing effector memory markers. This indicates that CD4 T cell differentiation status differentially affects LRA effectiveness.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no conflicts of interest to report.
Figures



Comment in
-
HIV Reservoir: Finding the Right Needles in a Needlestack.Cell Host Microbe. 2016 Sep 14;20(3):280-282. doi: 10.1016/j.chom.2016.08.011. Cell Host Microbe. 2016. PMID: 27631698
References
-
- Arganaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. J Biol Chem. 2003;278:33912–33919. - PubMed
-
- Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. PD-1 and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

