Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses
- PMID: 27545046
- PMCID: PMC7105029
- DOI: 10.1016/j.chom.2016.07.008
Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses
Abstract
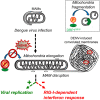
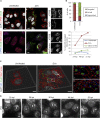
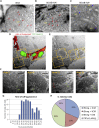
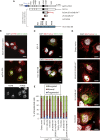
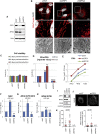
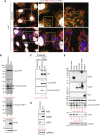
With no antiviral drugs or widely available vaccines, Dengue virus (DENV) constitutes a public health concern. DENV replicates at ER-derived cytoplasmic structures that include substructures called convoluted membranes (CMs); however, the purpose of these membrane alterations remains unclear. We determine that DENV nonstructural protein (NS)4B, a promising drug target with unknown function, associates with mitochondrial proteins and alters mitochondria morphology to promote infection. During infection, NS4B induces elongation of mitochondria, which physically contact CMs. This restructuring compromises the integrity of mitochondria-associated membranes, sites of ER-mitochondria interface critical for innate immune signaling. The spatio-temporal parameters of CM biogenesis and mitochondria elongation are linked to loss of activation of the fission factor Dynamin-Related Protein-1. Mitochondria elongation promotes DENV replication and alleviates RIG-I-dependent activation of interferon responses. As Zika virus infection induces similar mitochondria elongation, this perturbation may protect DENV and related viruses from innate immunity and create a favorable replicative environment.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Viral infection: Dengue as mitochondrial landscapers.Nat Rev Microbiol. 2016 Oct;14(10):603. doi: 10.1038/nrmicro.2016.132. Epub 2016 Aug 30. Nat Rev Microbiol. 2016. PMID: 27573576 No abstract available.
References
-
- Acosta E.G., Kumar A., Bartenschlager R. Revisiting dengue virus-host cell interaction: new insights into molecular and cellular virology. Adv. Virus Res. 2014;88:1–109. - PubMed
-
- Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012;46:265–287. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

