Building on the Ccr4-Not architecture
- PMID: 27545501
- PMCID: PMC5108432
- DOI: 10.1002/bies.201600051
Building on the Ccr4-Not architecture
Abstract
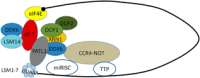
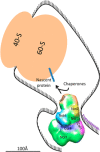
In a recent issue of Nature Communications Ukleja and co-workers reported a cryo-EM 3D reconstruction of the Ccr4-Not complex from Schizosaccharomyces pombe with an immunolocalization of the different subunits. The newly gained architectural knowledge provides cues to apprehend the functional diversity of this major eukaryotic regulator. Indeed, in the cytoplasm alone, Ccr4-Not regulates translational repression, decapping and deadenylation, and the Not module additionally plays a positive role in translation. The spatial distribution of the subunits within the structure is compatible with a model proposing that the Ccr4-Not complex interacts with the 5' and 3' ends of target mRNAs, allowing different functional modules of the complex to act at different stages of the translation process, possibly within a circular constellation of the mRNA. This work opens new avenues, and reveals important gaps in our understanding regarding structure and mode of function of the Ccr4-Not complex that need to be addressed in the future.
Keywords: Ccr4-Not complex; architecture; deadenylation; mRNA circularization; mRNA decay; protein folding; subunit localization.
© 2016 The Authors. Bioessays published by WILEY Periodicals, Inc.
Figures
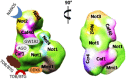
Similar articles
-
The architecture of the Schizosaccharomyces pombe CCR4-NOT complex.Nat Commun. 2016 Jan 25;7:10433. doi: 10.1038/ncomms10433. Nat Commun. 2016. PMID: 26804377 Free PMC article.
-
The enzyme activities of Caf1 and Ccr4 are both required for deadenylation by the human Ccr4-Not nuclease module.Biochem J. 2015 Jul 1;469(1):169-76. doi: 10.1042/BJ20150304. Epub 2015 May 6. Biochem J. 2015. PMID: 25944446 Free PMC article.
-
The Not3/5 subunit of the Ccr4-Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells.Cell Signal. 2013 Apr;25(4):743-51. doi: 10.1016/j.cellsig.2012.12.018. Epub 2012 Dec 29. Cell Signal. 2013. PMID: 23280189 Review.
-
RNF219 regulates CCR4-NOT function in mRNA translation and deadenylation.Sci Rep. 2022 Jun 3;12(1):9288. doi: 10.1038/s41598-022-13309-8. Sci Rep. 2022. PMID: 35660762 Free PMC article.
-
The Ccr4-Not complex is a key regulator of eukaryotic gene expression.Wiley Interdiscip Rev RNA. 2016 Jul;7(4):438-54. doi: 10.1002/wrna.1332. Epub 2016 Jan 29. Wiley Interdiscip Rev RNA. 2016. PMID: 26821858 Free PMC article. Review.
Cited by
-
CNOT6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte.EMBO J. 2018 Dec 14;37(24):e99333. doi: 10.15252/embj.201899333. Epub 2018 Nov 26. EMBO J. 2018. PMID: 30478191 Free PMC article.
-
The CNOT4 Subunit of the CCR4-NOT Complex is Involved in mRNA Degradation, Efficient DNA Damage Repair, and XY Chromosome Crossover during Male Germ Cell Meiosis.Adv Sci (Weinh). 2021 Mar 16;8(10):2003636. doi: 10.1002/advs.202003636. eCollection 2021 May. Adv Sci (Weinh). 2021. PMID: 34026442 Free PMC article.
-
Uncovering a novel function of the CCR4-NOT complex in phytochrome A-mediated light signalling in plants.Elife. 2021 Mar 30;10:e63697. doi: 10.7554/eLife.63697. Elife. 2021. PMID: 33783355 Free PMC article.
-
The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.Cell Rep. 2023 Jan 31;42(1):111902. doi: 10.1016/j.celrep.2022.111902. Epub 2022 Dec 30. Cell Rep. 2023. PMID: 36586408 Free PMC article.
References
-
- Villanyi Z, Collart MA. 2015. Ccr4‐Not is at the core of the eukaryotic gene expression circuitry. Biochem Soc Trans 43: 1253–8. - PubMed
-
- Chapat C, Corbo L. 2014. Novel roles of the CCR4‐NOT complex. Wiley Interdiscip Rev RNA 5: 883–901. - PubMed
-
- Collart MA, Panasenko OO. 2012. The Ccr4‐Not complex. Gene 492: 42–53. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

