Biallelic Variants in UBA5 Link Dysfunctional UFM1 Ubiquitin-like Modifier Pathway to Severe Infantile-Onset Encephalopathy
- PMID: 27545674
- PMCID: PMC5010641
- DOI: 10.1016/j.ajhg.2016.06.020
Biallelic Variants in UBA5 Link Dysfunctional UFM1 Ubiquitin-like Modifier Pathway to Severe Infantile-Onset Encephalopathy
Abstract
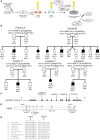
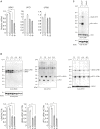
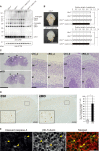
The ubiquitin fold modifier 1 (UFM1) cascade is a recently identified evolutionarily conserved ubiquitin-like modification system whose function and link to human disease have remained largely uncharacterized. By using exome sequencing in Finnish individuals with severe epileptic syndromes, we identified pathogenic compound heterozygous variants in UBA5, encoding an activating enzyme for UFM1, in two unrelated families. Two additional individuals with biallelic UBA5 variants were identified from the UK-based Deciphering Developmental Disorders study and one from the Northern Finland Intellectual Disability cohort. The affected individuals (n = 9) presented in early infancy with severe irritability, followed by dystonia and stagnation of development. Furthermore, the majority of individuals display postnatal microcephaly and epilepsy and develop spasticity. The affected individuals were compound heterozygous for a missense substitution, c.1111G>A (p.Ala371Thr; allele frequency of 0.28% in Europeans), and a nonsense variant or c.164G>A that encodes an amino acid substitution p.Arg55His, but also affects splicing by facilitating exon 2 skipping, thus also being in effect a loss-of-function allele. Using an in vitro thioester formation assay and cellular analyses, we show that the p.Ala371Thr variant is hypomorphic with attenuated ability to transfer the activated UFM1 to UFC1. Finally, we show that the CNS-specific knockout of Ufm1 in mice causes neonatal death accompanied by microcephaly and apoptosis in specific neurons, further suggesting that the UFM1 system is essential for CNS development and function. Taken together, our data imply that the combination of a hypomorphic p.Ala371Thr variant in trans with a loss-of-function allele in UBA5 underlies a severe infantile-onset encephalopathy.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Biallelic Variants in UBA5 Reveal that Disruption of the UFM1 Cascade Can Result in Early-Onset Encephalopathy.Am J Hum Genet. 2016 Sep 1;99(3):695-703. doi: 10.1016/j.ajhg.2016.06.030. Epub 2016 Aug 18. Am J Hum Genet. 2016. PMID: 27545681 Free PMC article.
-
Compound heterozygous mutations in UBA5 causing early-onset epileptic encephalopathy in two sisters.BMC Med Genet. 2017 Oct 2;18(1):103. doi: 10.1186/s12881-017-0466-8. BMC Med Genet. 2017. PMID: 28965491 Free PMC article.
-
A description of novel variants and review of phenotypic spectrum in UBA5-related early epileptic encephalopathy.Cold Spring Harb Mol Case Stud. 2021 Jun 11;7(3):a005827. doi: 10.1101/mcs.a005827. Print 2021 Jun. Cold Spring Harb Mol Case Stud. 2021. PMID: 33811063 Free PMC article.
-
UFMylation: A Unique & Fashionable Modification for Life.Genomics Proteomics Bioinformatics. 2016 Jun;14(3):140-146. doi: 10.1016/j.gpb.2016.04.001. Epub 2016 May 20. Genomics Proteomics Bioinformatics. 2016. PMID: 27212118 Free PMC article. Review.
-
Emerging role of protein modification by UFM1 in cancer.Biochem Biophys Res Commun. 2022 Dec 10;633:61-63. doi: 10.1016/j.bbrc.2022.08.093. Biochem Biophys Res Commun. 2022. PMID: 36344165 Review.
Cited by
-
The emerging roles of UFMylation in the modulation of immune responses.Clin Transl Med. 2024 Sep;14(9):e70019. doi: 10.1002/ctm2.70019. Clin Transl Med. 2024. PMID: 39259506 Free PMC article. Review.
-
The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.Nat Commun. 2022 Dec 21;13(1):7857. doi: 10.1038/s41467-022-35501-0. Nat Commun. 2022. PMID: 36543799 Free PMC article.
-
Mechanistic insights into the roles of the UFM1 E3 ligase complex in ufmylation and ribosome-associated protein quality control.Sci Adv. 2023 Aug 18;9(33):eadh3635. doi: 10.1126/sciadv.adh3635. Epub 2023 Aug 18. Sci Adv. 2023. PMID: 37595036 Free PMC article.
-
A 2020 View on the Genetics of Developmental and Epileptic Encephalopathies.Epilepsy Curr. 2020 Mar;20(2):90-96. doi: 10.1177/1535759720906118. Epub 2020 Mar 13. Epilepsy Curr. 2020. PMID: 32166973 Free PMC article.
-
Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development.Brain. 2018 Jul 1;141(7):1934-1945. doi: 10.1093/brain/awy135. Brain. 2018. PMID: 29868776 Free PMC article.
References
-
- van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. - PubMed
-
- Kang S.H., Kim G.R., Seong M., Baek S.H., Seol J.H., Bang O.S., Ovaa H., Tatsumi K., Komatsu M., Tanaka K., Chung C.H. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 2007;282:5256–5262. - PubMed
-
- Yoo H.M., Kang S.H., Kim J.Y., Lee J.E., Seong M.W., Lee S.W., Ka S.H., Sou Y.S., Komatsu M., Tanaka K. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell. 2014;56:261–274. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

