Biallelic Variants in UBA5 Reveal that Disruption of the UFM1 Cascade Can Result in Early-Onset Encephalopathy
- PMID: 27545681
- PMCID: PMC5011045
- DOI: 10.1016/j.ajhg.2016.06.030
Biallelic Variants in UBA5 Reveal that Disruption of the UFM1 Cascade Can Result in Early-Onset Encephalopathy
Abstract
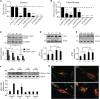
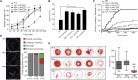
Via whole-exome sequencing, we identified rare autosomal-recessive variants in UBA5 in five children from four unrelated families affected with a similar pattern of severe intellectual deficiency, microcephaly, movement disorders, and/or early-onset intractable epilepsy. UBA5 encodes the E1-activating enzyme of ubiquitin-fold modifier 1 (UFM1), a recently identified ubiquitin-like protein. Biochemical studies of mutant UBA5 proteins and studies in fibroblasts from affected individuals revealed that UBA5 mutations impair the process of ufmylation, resulting in an abnormal endoplasmic reticulum structure. In Caenorhabditis elegans, knockout of uba-5 and of human orthologous genes in the UFM1 cascade alter cholinergic, but not glutamatergic, neurotransmission. In addition, uba5 silencing in zebrafish decreased motility while inducing abnormal movements suggestive of seizures. These clinical, biochemical, and experimental findings support our finding of UBA5 mutations as a pathophysiological cause for early-onset encephalopathies due to abnormal protein ufmylation.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development.Brain. 2018 Jul 1;141(7):1934-1945. doi: 10.1093/brain/awy135. Brain. 2018. PMID: 29868776 Free PMC article.
-
Biallelic Variants in UBA5 Link Dysfunctional UFM1 Ubiquitin-like Modifier Pathway to Severe Infantile-Onset Encephalopathy.Am J Hum Genet. 2016 Sep 1;99(3):683-694. doi: 10.1016/j.ajhg.2016.06.020. Epub 2016 Aug 18. Am J Hum Genet. 2016. PMID: 27545674 Free PMC article.
-
Allelic strengths of encephalopathy-associated UBA5 variants correlate between in vivo and in vitro assays.Elife. 2023 Dec 11;12:RP89891. doi: 10.7554/eLife.89891. Elife. 2023. PMID: 38079206 Free PMC article.
-
Emerging role of protein modification by UFM1 in cancer.Biochem Biophys Res Commun. 2022 Dec 10;633:61-63. doi: 10.1016/j.bbrc.2022.08.093. Biochem Biophys Res Commun. 2022. PMID: 36344165 Review.
-
UFMylation: A Unique & Fashionable Modification for Life.Genomics Proteomics Bioinformatics. 2016 Jun;14(3):140-146. doi: 10.1016/j.gpb.2016.04.001. Epub 2016 May 20. Genomics Proteomics Bioinformatics. 2016. PMID: 27212118 Free PMC article. Review.
Cited by
-
A novel compound heterozygous mutation of UFC1 in a patient with neurodevelopmental disorder.Genes Genomics. 2024 Sep;46(9):1037-1043. doi: 10.1007/s13258-024-01543-5. Epub 2024 Jul 29. Genes Genomics. 2024. PMID: 39078589
-
The UFMylation System in Proteostasis and Beyond.Trends Cell Biol. 2019 Dec;29(12):974-986. doi: 10.1016/j.tcb.2019.09.005. Epub 2019 Nov 6. Trends Cell Biol. 2019. PMID: 31703843 Free PMC article. Review.
-
Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development.Brain. 2018 Jul 1;141(7):1934-1945. doi: 10.1093/brain/awy135. Brain. 2018. PMID: 29868776 Free PMC article.
-
Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR.Nat Commun. 2019 Mar 6;10(1):1084. doi: 10.1038/s41467-019-08908-5. Nat Commun. 2019. PMID: 30842412 Free PMC article.
-
Retrotransposon insertions associated with risk of neurologic and psychiatric diseases.EMBO Rep. 2023 Jan 9;24(1):e55197. doi: 10.15252/embr.202255197. Epub 2022 Nov 11. EMBO Rep. 2023. PMID: 36367221 Free PMC article.
References
-
- van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

