High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA
- PMID: 27545715
- PMCID: PMC6640135
- DOI: 10.1016/j.neuron.2016.07.036
High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA
Abstract
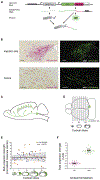
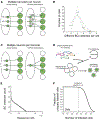
Neurons transmit information to distant brain regions via long-range axonal projections. In the mouse, area-to-area connections have only been systematically mapped using bulk labeling techniques, which obscure the diverse projections of intermingled single neurons. Here we describe MAPseq (Multiplexed Analysis of Projections by Sequencing), a technique that can map the projections of thousands or even millions of single neurons by labeling large sets of neurons with random RNA sequences ("barcodes"). Axons are filled with barcode mRNA, each putative projection area is dissected, and the barcode mRNA is extracted and sequenced. Applying MAPseq to the locus coeruleus (LC), we find that individual LC neurons have preferred cortical targets. By recasting neuroanatomy, which is traditionally viewed as a problem of microscopy, as a problem of sequencing, MAPseq harnesses advances in sequencing technology to permit high-throughput interrogation of brain circuits.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interests
The authors declare no conflict of interests.
Figures





References
-
- Aston-Jones G, and Cohen JD (2005). An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Annu. Rev. Neurosci. 28, 403–450. - PubMed
-
- Cetin A, Komai S, Eliava M, Seeburg PH, and Osten P (2007). Stereotaxic gene delivery in the rodent brain. Nat Protoc 1, 3166–3173. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

