Synthetic Biology Platform for Sensing and Integrating Endogenous Transcriptional Inputs in Mammalian Cells
- PMID: 27545896
- PMCID: PMC5009115
- DOI: 10.1016/j.celrep.2016.07.061
Synthetic Biology Platform for Sensing and Integrating Endogenous Transcriptional Inputs in Mammalian Cells
Abstract
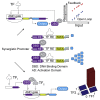
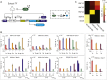
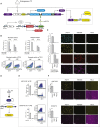
One of the goals of synthetic biology is to develop programmable artificial gene networks that can transduce multiple endogenous molecular cues to precisely control cell behavior. Realizing this vision requires interfacing natural molecular inputs with synthetic components that generate functional molecular outputs. Interfacing synthetic circuits with endogenous mammalian transcription factors has been particularly difficult. Here, we describe a systematic approach that enables integration and transduction of multiple mammalian transcription factor inputs by a synthetic network. The approach is facilitated by a proportional amplifier sensor based on synergistic positive autoregulation. The circuits efficiently transduce endogenous transcription factor levels into RNAi, transcriptional transactivation, and site-specific recombination. They also enable AND logic between pairs of arbitrary transcription factors. The results establish a framework for developing synthetic gene networks that interface with cellular processes through transcriptional regulators.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
MicroRNA circuits for transcriptional logic.Methods Mol Biol. 2012;813:169-86. doi: 10.1007/978-1-61779-412-4_10. Methods Mol Biol. 2012. PMID: 22083742
-
Engineering molecular circuits using synthetic biology in mammalian cells.Annu Rev Chem Biomol Eng. 2012;3:209-34. doi: 10.1146/annurev-chembioeng-061010-114145. Epub 2012 Mar 29. Annu Rev Chem Biomol Eng. 2012. PMID: 22468602 Review.
-
Programmable genetic circuits for pathway engineering.Curr Opin Biotechnol. 2015 Dec;36:115-21. doi: 10.1016/j.copbio.2015.08.007. Epub 2015 Aug 29. Curr Opin Biotechnol. 2015. PMID: 26322736 Review.
-
Synthetic biology with RNA: progress report.Curr Opin Chem Biol. 2012 Aug;16(3-4):278-84. doi: 10.1016/j.cbpa.2012.05.192. Epub 2012 Jun 6. Curr Opin Chem Biol. 2012. PMID: 22676891 Review.
-
Mammalian Synthetic Biology: Engineering Biological Systems.Annu Rev Biomed Eng. 2017 Jun 21;19:249-277. doi: 10.1146/annurev-bioeng-071516-044649. Annu Rev Biomed Eng. 2017. PMID: 28633563 Review.
Cited by
-
De novo design of protein logic gates.Science. 2020 Apr 3;368(6486):78-84. doi: 10.1126/science.aay2790. Science. 2020. PMID: 32241946 Free PMC article.
-
Rational gRNA design based on transcription factor binding data.Synth Biol (Oxf). 2021 Jul 27;6(1):ysab014. doi: 10.1093/synbio/ysab014. eCollection 2021. Synth Biol (Oxf). 2021. PMID: 34712839 Free PMC article.
-
CAGE: Chromatin Analogous Gene Expression.ACS Synth Biol. 2017 Oct 20;6(10):1800-1806. doi: 10.1021/acssynbio.7b00045. Epub 2017 Jun 30. ACS Synth Biol. 2017. PMID: 28657718 Free PMC article.
-
Extending dynamic and operational range of the biosensor responding to l-carnitine by directed evolution.Synth Syst Biotechnol. 2025 Apr 22;10(3):897-906. doi: 10.1016/j.synbio.2025.04.012. eCollection 2025 Sep. Synth Syst Biotechnol. 2025. PMID: 40386442 Free PMC article.
-
Oncolytic adenovirus programmed by synthetic gene circuit for cancer immunotherapy.Nat Commun. 2019 Oct 22;10(1):4801. doi: 10.1038/s41467-019-12794-2. Nat Commun. 2019. PMID: 31641136 Free PMC article.
References
-
- Acar M., Becskei A., van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. - PubMed
-
- Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. - PubMed
-
- Ausländer S., Ausländer D., Müller M., Wieland M., Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

