Members of the Hsp70 Family Recognize Distinct Types of Sequences to Execute ER Quality Control
- PMID: 27546788
- PMCID: PMC5010498
- DOI: 10.1016/j.molcel.2016.07.012
Members of the Hsp70 Family Recognize Distinct Types of Sequences to Execute ER Quality Control
Abstract
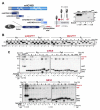
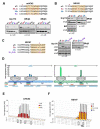
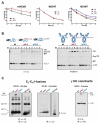
Protein maturation in the endoplasmic reticulum is controlled by multiple chaperones, but how they recognize and determine the fate of their clients remains unclear. We developed an in vivo peptide library covering substrates of the ER Hsp70 system: BiP, Grp170, and three of BiP's DnaJ-family co-factors (ERdj3, ERdj4, and ERdj5). In vivo binding studies revealed that sites for pro-folding chaperones BiP and ERdj3 were frequent and dispersed throughout the clients, whereas Grp170, ERdj4, and ERdj5 specifically recognized a distinct type of rarer sequence with a high predicted aggregation potential. Mutational analyses provided insights into sequence recognition characteristics for these pro-degradation chaperones, which could be readily introduced or disrupted, allowing the consequences for client fates to be determined. Our data reveal unanticipated diversity in recognition sequences for chaperones; establish a sequence-encoded interplay between protein folding, aggregation, and degradation; and highlight the ability of clients to co-evolve with chaperones, ensuring quality control.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Division of Labor: ER-Resident BiP Co-Chaperones Match Substrates to Fates Based on Specific Binding Sequences.Mol Cell. 2016 Sep 1;63(5):721-3. doi: 10.1016/j.molcel.2016.08.017. Mol Cell. 2016. PMID: 27588598 Free PMC article.
References
-
- Ahmed AB, Kajava AV. Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. FEBS Lett. 2013;587:1089–1095. - PubMed
-
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

