Single-Molecule FISH Reveals Non-selective Packaging of Rift Valley Fever Virus Genome Segments
- PMID: 27548280
- PMCID: PMC4993503
- DOI: 10.1371/journal.ppat.1005800
Single-Molecule FISH Reveals Non-selective Packaging of Rift Valley Fever Virus Genome Segments
Abstract
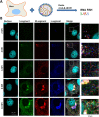
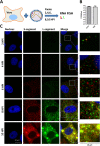
The bunyavirus genome comprises a small (S), medium (M), and large (L) RNA segment of negative polarity. Although genome segmentation confers evolutionary advantages by enabling genome reassortment events with related viruses, genome segmentation also complicates genome replication and packaging. Accumulating evidence suggests that genomes of viruses with eight or more genome segments are incorporated into virions by highly selective processes. Remarkably, little is known about the genome packaging process of the tri-segmented bunyaviruses. Here, we evaluated, by single-molecule RNA fluorescence in situ hybridization (FISH), the intracellular spatio-temporal distribution and replication kinetics of the Rift Valley fever virus (RVFV) genome and determined the segment composition of mature virions. The results reveal that the RVFV genome segments start to replicate near the site of infection before spreading and replicating throughout the cytoplasm followed by translocation to the virion assembly site at the Golgi network. Despite the average intracellular S, M and L genome segments approached a 1:1:1 ratio, major differences in genome segment ratios were observed among cells. We also observed a significant amount of cells lacking evidence of M-segment replication. Analysis of two-segmented replicons and four-segmented viruses subsequently confirmed the previous notion that Golgi recruitment is mediated by the Gn glycoprotein. The absence of colocalization of the different segments in the cytoplasm and the successful rescue of a tri-segmented variant with a codon shuffled M-segment suggested that inter-segment interactions are unlikely to drive the copackaging of the different segments into a single virion. The latter was confirmed by direct visualization of RNPs inside mature virions which showed that the majority of virions lack one or more genome segments. Altogether, this study suggests that RVFV genome packaging is a non-selective process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Flick R, Bouloy M (2005) Rift Valley fever virus. Curr Mol Med 5: 827–834. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

