Minimum information required for a DMET experiment reporting
- PMID: 27548815
- PMCID: PMC6123892
- DOI: 10.2217/pgs-2016-0015
Minimum information required for a DMET experiment reporting
Abstract
Aim: To provide pharmacogenomics reporting guidelines, the information and tools required for reporting to public omic databases.
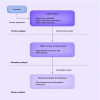
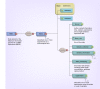
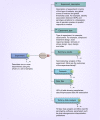
Material & methods: For effective DMET data interpretation, sharing, interoperability, reproducibility and reporting, we propose the Minimum Information required for a DMET Experiment (MIDE) reporting.
Results: MIDE provides reporting guidelines and describes the information required for reporting, data storage and data sharing in the form of XML.
Conclusion: The MIDE guidelines will benefit the scientific community with pharmacogenomics experiments, including reporting pharmacogenomics data from other technology platforms, with the tools that will ease and automate the generation of such reports using the standardized MIDE XML schema, facilitating the sharing, dissemination, reanalysis of datasets through accessible and transparent pharmacogenomics data reporting.
Keywords: DMET; bioinformatics; minimum information requirement guidelines; personalized genomics; personalized medicine; pharmacogenomics; standardization.
Conflict of interest statement
The CPGR is supported through institutional funding from TIA (Technology Innovation Agency) and the DST (Department of Science and Technology). This work is partially funded by NIH Common Fund Award/NHGRI Grant Number U41HG006941 through H3AbioNet project. M Macek was supported by NF-CZ11-PDP-3-003-2014, 00064203 and COST LD14073. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures
Similar articles
-
DMET-analyzer: automatic analysis of Affymetrix DMET data.BMC Bioinformatics. 2012 Oct 5;13:258. doi: 10.1186/1471-2105-13-258. BMC Bioinformatics. 2012. PMID: 23035929 Free PMC article.
-
DMET-Miner: Efficient discovery of association rules from pharmacogenomic data.J Biomed Inform. 2015 Aug;56:273-83. doi: 10.1016/j.jbi.2015.06.005. Epub 2015 Jun 16. J Biomed Inform. 2015. PMID: 26092773
-
DMET microarray technology for pharmacogenomics-based personalized medicine.Methods Mol Biol. 2010;632:99-124. doi: 10.1007/978-1-60761-663-4_7. Methods Mol Biol. 2010. PMID: 20217574
-
DMETTM Genotyping: Tools for Biomarkers Discovery in the Era of Precision Medicine.High Throughput. 2020 Mar 29;9(2):8. doi: 10.3390/ht9020008. High Throughput. 2020. PMID: 32235355 Free PMC article. Review.
-
The minimum information required for reporting a molecular interaction experiment (MIMIx).Nat Biotechnol. 2007 Aug;25(8):894-8. doi: 10.1038/nbt1324. Nat Biotechnol. 2007. PMID: 17687370 Review.
Cited by
-
Tolerogenic dendritic cell reporting: Has a minimum information model made a difference?PeerJ. 2023 May 31;11:e15352. doi: 10.7717/peerj.15352. eCollection 2023. PeerJ. 2023. PMID: 37273539 Free PMC article.
-
Implementing the FAIR Data Principles in precision oncology: review of supporting initiatives.Brief Bioinform. 2020 May 21;21(3):936-945. doi: 10.1093/bib/bbz044. Brief Bioinform. 2020. PMID: 31263868 Free PMC article. Review.
-
Ten simple rules for providing effective bioinformatics research support.PLoS Comput Biol. 2020 Mar 26;16(3):e1007531. doi: 10.1371/journal.pcbi.1007531. eCollection 2020 Mar. PLoS Comput Biol. 2020. PMID: 32214318 Free PMC article.
References
-
- Thomas FJ, McLeod HL, Watters JW. Pharmacogenomics: the influence of genomic variation on drug response. Curr. Top. Med. Chem. 2004;4(13):1399–1409. - PubMed
-
• Highlights essential integration of both pharmacogenomic and clinical data can provide powerful predictive tools for stratifying responders from nonresponders and identifying patients at increased risk for toxicity.
-
- Watson RG, McLeod HL. Pharmacogenomic contribution to drug response. Cancer J. 2011;17(2):80–88. - PubMed
-
- Pirmohamed M. Drug metabolism. Medicine. 2008;36(7):355–359.
-
- Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu. Rev. Genomics Hum. Genet. 2006;7:223–245. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
