Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa
- PMID: 27549218
- PMCID: PMC4994406
- DOI: 10.1186/s12870-016-0870-9
Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa
Abstract
Background: Flavonoids are secondary metabolites that play significant roles in plant cells. In particular, polymethoxy flavonoids (PMFs), including nobiletin, have been reported to exhibit various health-supporting properties such as anticancer, anti-inflammatory, and anti-pathogenic properties. However, it is difficult to utilize PMFs for medicinal and dietary use because plant cells contain small amounts of these compounds. Biosynthesis of PMFs in plant cells is carried out by the methylation of hydroxyl groups of flavonoids by O-methyltransferases (FOMT), and many kinds of FOMTs with different levels of substrate specificity and regioselectivity are cooperatively involved in this biosynthesis.
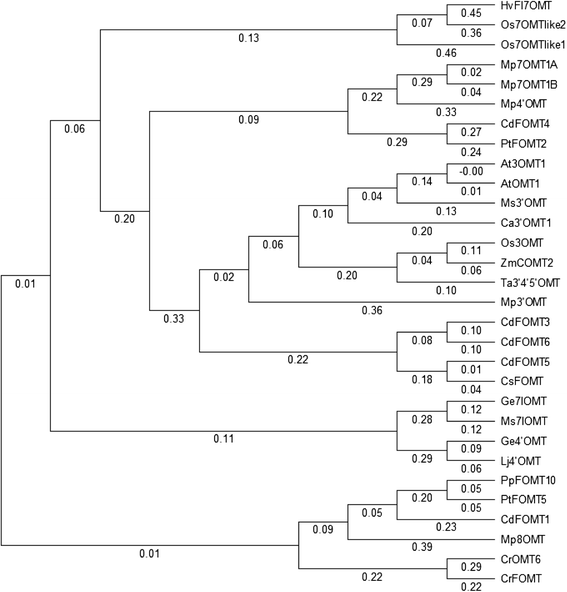
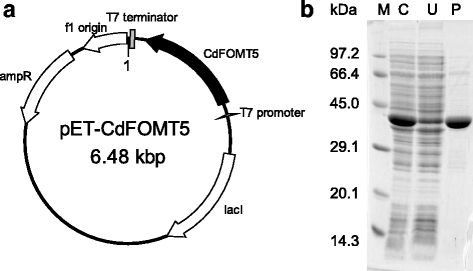
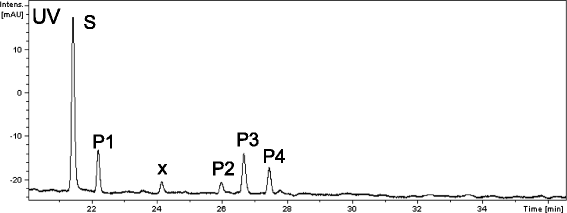
Results: In this study, we isolated five genes encoding FOMT (CdFOMT1, 3, 4, 5, and 6) from Citrus depressa, which is known to accumulate nobiletin in the peels of its fruits. The genes encoded Mg(2+)-independent O-methyltransferases and showed high amino acid sequence similarity (60-95 %) with higher plant flavonoid O-methyltransferases. One of these genes is CdFOMT5, which was successfully expressed as a soluble homodimer enzyme in Escherichia coli. The molecular mass of the recombinant CdFOMT5 subunit was 42.0 kDa including a 6× histidine tag. The enzyme exhibited O-methyltransferase activity for quercetin, naringenin, (-)-epicatechin, and equol using S-adenosyl-L-methionine (SAM) as a methyl donor, and its optimal pH and temperature were pH 7.0 and 45 °C, respectively. The recombinant CdFOMT5 demonstrated methylation activity for the 3-, 5-, 6-, and 7-hydroxyl groups of flavones, and 3,3',5,7-tetra-O-methylated quercetin was synthesized from quercetin as a final product of the whole cell reaction system. Thus, CdFOMT5 is a O-methyltransferase possessing a broad range of substrate specificity and regioselectivity for flavonoids.
Conclusions: Five FOMT genes were isolated from C. depressa, and their nucleotide sequences were determined. CdFOMT5 was successfully expressed in E. coli cells, and the enzymatic properties of the recombinant protein were characterized. Recombinant CdFOMT5 indicated O-methyltransferase activity for many flavonoids and a broad regioselectivity for quercetin as a substrate. Whole-cell biocatalysis using CdFOMT5 expressed in E. coli cells was performed using quercetin as a substrate, and 3,3',5,7-tetramethylated quercetin was obtained as the final product.
Keywords: Citrus depressa; O-methyltransferase; S-adenosyl-L-methionine dependent; flavonoid; nobiletin.
Figures



Similar articles
-
Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds.Arch Biochem Biophys. 1998 Mar 15;351(2):243-9. doi: 10.1006/abbi.1997.0554. Arch Biochem Biophys. 1998. PMID: 9514654
-
Regiospecific flavonoid 7-O-methylation with Streptomyces avermitilis O-methyltransferase expressed in Escherichia coli.J Agric Food Chem. 2006 Feb 8;54(3):823-8. doi: 10.1021/jf0522715. J Agric Food Chem. 2006. PMID: 16448189
-
Flavonoid methylation: a novel 4'-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases.Phytochemistry. 2004 Apr;65(8):1085-94. doi: 10.1016/j.phytochem.2004.02.010. Phytochemistry. 2004. PMID: 15110688
-
Diversity and regioselectivity of O-methyltransferases catalyzing the formation of O-methylated flavonoids.Crit Rev Biotechnol. 2024 Sep;44(6):1203-1225. doi: 10.1080/07388551.2023.2280755. Epub 2023 Nov 30. Crit Rev Biotechnol. 2024. PMID: 38035668 Review.
-
Diversification of Chemical Structures of Methoxylated Flavonoids and Genes Encoding Flavonoid-O-Methyltransferases.Plants (Basel). 2022 Feb 21;11(4):564. doi: 10.3390/plants11040564. Plants (Basel). 2022. PMID: 35214897 Free PMC article. Review.
Cited by
-
Differences in flavonoid pathway metabolites and transcripts affect yellow petal colouration in the aquatic plant Nelumbo nucifera.BMC Plant Biol. 2019 Jun 24;19(1):277. doi: 10.1186/s12870-019-1886-8. BMC Plant Biol. 2019. PMID: 31234776 Free PMC article.
-
One-Pot Biocatalytic In Vivo Methylation-Hydroamination of Bioderived Lignin Monomers to Generate a Key Precursor to L-DOPA.Angew Chem Weinheim Bergstr Ger. 2022 Feb 14;134(8):e202112855. doi: 10.1002/ange.202112855. Epub 2022 Jan 10. Angew Chem Weinheim Bergstr Ger. 2022. PMID: 38505118 Free PMC article.
-
Expression and functional analysis of the nobiletin biosynthesis-related gene CitOMT in citrus fruit.Sci Rep. 2020 Sep 17;10(1):15288. doi: 10.1038/s41598-020-72277-z. Sci Rep. 2020. PMID: 32943728 Free PMC article.
-
A multifunctional true caffeoyl coenzyme A O-methyltransferase enzyme participates in the biosynthesis of polymethoxylated flavones in citrus.Plant Physiol. 2023 Jul 3;192(3):2049-2066. doi: 10.1093/plphys/kiad249. Plant Physiol. 2023. PMID: 37086474 Free PMC article.
-
Genome-wide identification and characterization of COMT gene family during the development of blueberry fruit.BMC Plant Biol. 2021 Jan 6;21(1):5. doi: 10.1186/s12870-020-02767-9. BMC Plant Biol. 2021. PMID: 33407129 Free PMC article.
References
-
- Koes RE, Quattrocchio F, Mol JNM. The flavonoid biosynthetic pathway in plants: Function and evolution. Bio Essays. 1994;16(2):123–132.
-
- Cook NC, Samman S. Flavonoids-Chemistry, metabolism, cardioprotective effects, and dietary sources. J Nutr Biochem. 1996;7(2):66–76. doi: 10.1016/0955-2863(95)00168-9. - DOI
-
- Peterson JMS, Dwyer J, Dsc RD. Flavonoids: Dietary occurrence and biochemical activity. Nutr Res. 1998;18(12):1995–2018. doi: 10.1016/S0271-5317(98)00169-9. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

