High-fat diet disrupts metabolism in two generations of rats in a parent-of-origin specific manner
- PMID: 27550193
- PMCID: PMC4994008
- DOI: 10.1038/srep31857
High-fat diet disrupts metabolism in two generations of rats in a parent-of-origin specific manner
Abstract
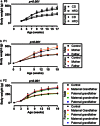
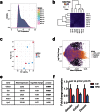
Experimental and epidemiological evidence demonstrate that ancestral diet might contribute towards offspring health. This suggests that nutrition may be able to modify genetic or epigenetic information carried by germ cells (GCs). To examine if a parental high fat diet (HFD) influences metabolic health in two generations of offspring, GC-eGFP Sprague Dawley rats were weaned onto HFD (45% fat) or Control Diet (CD; 10% fat). At 19 weeks, founders (F0) were bred with controls, establishing the F1 generation. HFD resulted in 9.7% and 14.7% increased weight gain in male and female F0 respectively. F1 offspring of HFD mothers and F1 daughters of HFD-fed fathers had increased weight gain compared to controls. F1 rats were bred with controls at 19 weeks to generate F2 offspring. F2 male offspring derived from HFD-fed maternal grandfathers exhibited increased adiposity, plasma leptin and luteinising hormone to testosterone ratio. Despite transmission via the founding male germline, we did not find significant changes in the F0 intra-testicular GC transcriptome. Thus, HFD consumption by maternal grandfathers results in a disrupted metabolic and reproductive hormone phenotype in grandsons in the absence of detectable changes in the intra-testicular GC transcriptome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Wang Y. C., McPherson K., Marsh T., Gortmaker S. L. & Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet 378, 815–825 (2011). - PubMed
-
- WHO. WHO| Obesity and overweight. WHO (2014). Available at: http://www.who.int/mediacentre/factsheets/fs311/en/. (Accessed: 21st May 2014).
-
- Patro B. et al. Maternal and Paternal Body Mass Index and Offspring Obesity: A Systematic Review. Ann. Nutr. Metab. 63, 32–41 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

