Mutations in TrkA Causing Congenital Insensitivity to Pain with Anhidrosis (CIPA) Induce Misfolding, Aggregation, and Mutation-dependent Neurodegeneration by Dysfunction of the Autophagic Flux
- PMID: 27551041
- PMCID: PMC5076807
- DOI: 10.1074/jbc.M116.722587
Mutations in TrkA Causing Congenital Insensitivity to Pain with Anhidrosis (CIPA) Induce Misfolding, Aggregation, and Mutation-dependent Neurodegeneration by Dysfunction of the Autophagic Flux
Abstract
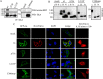
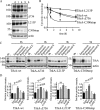
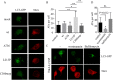
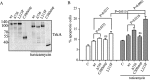
Congenital insensitivity to pain with anhidrosis (CIPA) is a rare autosomal recessive disorder characterized by insensitivity to noxious stimuli and variable intellectual disability (ID) due to mutations in the NTRK1 gene encoding the NGF receptor TrkA. To get an insight in the effect of NTRK1 mutations in the cognitive phenotype we biochemically characterized three TrkA mutations identified in children diagnosed of CIPA with variable ID. These mutations are located in different domains of the protein; L213P in the extracellular domain, Δ736 in the kinase domain, and C300stop in the extracellular domain, a new mutation causing CIPA diagnosed in a Spanish teenager. We found that TrkA mutations induce misfolding, retention in the endoplasmic reticulum (ER), and aggregation in a mutation-dependent manner. The distinct mutations are degraded with a different kinetics by different ER quality control mechanisms; although C300stop is rapidly disposed by autophagy, Δ736 degradation is sensitive to the proteasome and to autophagy inhibitors, and L213P is a long-lived protein refractory to degradation. In addition L213P enhances the formation of autophagic vesicles triggering an increase in the autophagic flux with deleterious consequences. Mouse cortical neurons expressing L213P showed the accumulation of LC3-GFP positive puncta and dystrophic neurites. Our data suggest that TrkA misfolding and aggregation induced by some CIPA mutations disrupt the autophagy homeostasis causing neurodegeneration. We propose that distinct disease-causing mutations of TrkA generate different levels of cell toxicity, which may provide an explanation of the variable intellectual disability observed in CIPA patients.
Keywords: ER quality control; TRK1-transforming tyrosine kinase protein (Trk-A); autophagic flux; autophagy; congenital insensitivity to pain with anhidrosis; neurodegeneration; protein aggregation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Congenital insensitivity to pain with anhidrosis (CIPA): effect of TRKA (NTRK1) missense mutations on autophosphorylation of the receptor tyrosine kinase for nerve growth factor.Hum Mol Genet. 2001 Feb 1;10(3):179-88. doi: 10.1093/hmg/10.3.179. Hum Mol Genet. 2001. PMID: 11159935
-
Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis.Nat Genet. 1996 Aug;13(4):485-8. doi: 10.1038/ng0896-485. Nat Genet. 1996. PMID: 8696348
-
Novel pathogenic mechanisms of congenital insensitivity to pain with anhidrosis genetic disorder unveiled by functional analysis of neurotrophic tyrosine receptor kinase type 1/nerve growth factor receptor mutations.J Biol Chem. 2002 Feb 22;277(8):6455-62. doi: 10.1074/jbc.M110016200. Epub 2001 Nov 21. J Biol Chem. 2002. PMID: 11719521
-
Genetics of congenital insensitivity to pain with anhidrosis (CIPA) or hereditary sensory and autonomic neuropathy type IV. Clinical, biological and molecular aspects of mutations in TRKA(NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor.Clin Auton Res. 2002 May;12 Suppl 1:I20-32. doi: 10.1007/s102860200016. Clin Auton Res. 2002. PMID: 12102460 Review.
-
Molecular basis of congenital insensitivity to pain with anhidrosis (CIPA): mutations and polymorphisms in TRKA (NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor.Hum Mutat. 2001 Dec;18(6):462-71. doi: 10.1002/humu.1224. Hum Mutat. 2001. PMID: 11748840 Review.
Cited by
-
NTRK1 knockdown induces mouse cognitive impairment and hippocampal neuronal damage through mitophagy suppression via inactivating the AMPK/ULK1/FUNDC1 pathway.Cell Death Discov. 2023 Oct 31;9(1):404. doi: 10.1038/s41420-023-01685-7. Cell Death Discov. 2023. PMID: 37907480 Free PMC article.
-
Autophagy as an Emerging Common Pathomechanism in Inherited Peripheral Neuropathies.Front Mol Neurosci. 2017 May 11;10:143. doi: 10.3389/fnmol.2017.00143. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28553203 Free PMC article. Review.
-
Targeting NTRK1 Enhances Immune Checkpoint Inhibitor Efficacy in NTRK1 Wild-Type Non-Small Cell Lung Cancer.Cancer Res. 2024 Dec 2;84(23):4002-4016. doi: 10.1158/0008-5472.CAN-24-0658. Cancer Res. 2024. PMID: 39250241 Free PMC article.
-
Unidirectional Crosstalk Between NTRK1 and IGF2 Drives ER Stress in Chronic Pain.Biomedicines. 2025 Jul 3;13(7):1632. doi: 10.3390/biomedicines13071632. Biomedicines. 2025. PMID: 40722704 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
References
-
- Indo Y. (2010) Nerve growth factor, pain, itch and inflammation: lessons from congenital insensitivity to pain with anhidrosis. Expert Rev. Neurother. 10, 1707–1724 - PubMed
-
- Indo Y. (2002) Genetics of congenital insensitivity to pain with anhidrosis (CIPA) or hereditary sensory and autonomic neuropathy type IV. Clinical, biological and molecular aspects of mutations in TRKA(NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor. Clin. Auton. Res. 12, I20–32 - PubMed
-
- Sztriha L., Lestringant G. G., Hertecant J., Frossard P. M., and Masouyé I. (2001) Congenital insensitivity to pain with anhidrosis. Pediatr. Neurol. 25, 63–66 - PubMed
-
- Indo Y., Tsuruta M., Hayashida Y., Karim M. A., Ohta K., Kawano T., Mitsubuchi H., Tonoki H., Awaya Y., and Matsuda I. (1996) Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat. Genet. 13, 485–488 - PubMed
-
- Lallemend F., and Ernfors P. (2012) Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 35, 373–381 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

