The Steroid Hormone 20-Hydroxyecdysone Promotes the Cytoplasmic Localization of Yorkie to Suppress Cell Proliferation and Induce Apoptosis
- PMID: 27551043
- PMCID: PMC5076844
- DOI: 10.1074/jbc.M116.719856
The Steroid Hormone 20-Hydroxyecdysone Promotes the Cytoplasmic Localization of Yorkie to Suppress Cell Proliferation and Induce Apoptosis
Abstract
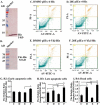
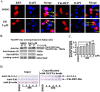
The transcriptional co-activator Yki (Yorkie), a member of the Hippo pathway, regulates cell proliferation or apoptosis, depending on its nuclear or cytoplasmic location. However, the upstream factors regulating the subcellular localization of Yki are unclear. We found that the steroid hormone 20-hydroxyecdysone (20E) induces phosphorylation of Yki, causing it to remain in the cytoplasm, where it promotes apoptosis in the midgut of the lepidopteran insect Helicoverpa armigera Yki is expressed in various tissues, with an increase in the epidermis and midgut during early metamorphic molting. Yki is localized mainly in the nucleus of feeding larval midgut cells but is mainly localized in the cytoplasm of metamorphic molting larval midgut cells. The knockdown of Yki in the feeding larvae promotes larval-pupal transition, midgut programmed cell death, and repressed IAP1 (inhibitor of apoptosis 1) expression. Knockdown of Yki in the epidermal cell line (HaEpi) induced increased activation of Caspase3/7. Overexpressed Yki in HaEpi cells was mainly localized in the nucleus and induced cell proliferation. 20E promotes the cytoplasmic localization of Yki, reducing the expression of the IAP1, resulting in apoptosis. 20E promotes cytoplasmic retention of Yki by increasing Yki phosphorylation levels and promoting the interaction between Yki and the adaptor protein 14-3-3-ϵ. This regulation of Yki suppresses cell proliferation and induces cell apoptosis.
Keywords: 20-hydroxyecdysone; Hippo pathway; Yorkie; apoptosis; cell proliferation; cytoplasmic localization; insect; steroid hormone.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Huang J., Wu S., Barrera J., Matthews K., and Pan D. (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

