Composition and Activity of the Non-canonical Gram-positive SecY2 Complex
- PMID: 27551046
- PMCID: PMC5076819
- DOI: 10.1074/jbc.M116.729806
Composition and Activity of the Non-canonical Gram-positive SecY2 Complex
Abstract
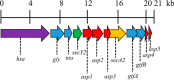
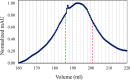
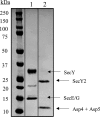
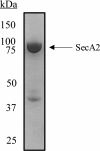
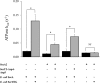
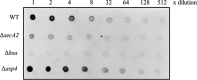
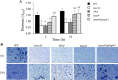
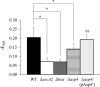
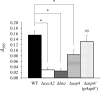
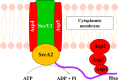
The accessory Sec system in Streptococcus gordonii DL1 is a specialized export system that transports a large serine-rich repeat protein, Hsa, to the bacterial surface. The system is composed of core proteins SecA2 and SecY2 and accessory Sec proteins Asp1-Asp5. Similar to canonical SecYEG, SecY2 forms a channel for translocation of the Hsa adhesin across the cytoplasmic membrane. Accessory Sec proteins Asp4 and Asp5 have been suggested to work alongside SecY2 to form the translocon, similar to the associated SecY, SecE, and SecG of the canonical system (SecYEG). To test this theory, S. gordonii secY2, asp4, and asp5 were co-expressed in Escherichia coli The resultant complex was subsequently purified, and its composition was confirmed by mass spectrometry to be SecY2-Asp4-Asp5. Like SecYEG, the non-canonical complex activates the ATPase activity of the SecA motor (SecA2). This study also shows that Asp4 and Asp5 are necessary for optimal adhesion of S. gordonii to glycoproteins gp340 and fibronectin, known Hsa binding partners, as well as for early stage biofilm formation. This work opens new avenues for understanding the structure and function of the accessory Sec system.
Keywords: ATPase; SecY2; Streptococcus gordonii; accessory Sec system; complex; membrane; secretion; translocon; transport.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Nyvad B., and Kilian M. (1990) Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24, 267–272 - PubMed
-
- Whittaker C. J., Klier C. M., and Kolenbrander P. E. (1996) Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50, 513–552 - PubMed
-
- Douglas C. W., Heath J., Hampton K. K., and Preston F. E. (1993) Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39, 179–182 - PubMed
-
- Jakubovics N. S., Kerrigan S. W., Nobbs A. H., Strömberg N., van Dolleweerd C. J., Cox D. M., Kelly C. G., and Jenkinson H. F. (2005) Functions of cell surface-A anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect. Immun. 73, 6629–6638 - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

