Severe Molecular Defects Exhibited by the R179H Mutation in Human Vascular Smooth Muscle α-Actin
- PMID: 27551047
- PMCID: PMC5076841
- DOI: 10.1074/jbc.M116.744011
Severe Molecular Defects Exhibited by the R179H Mutation in Human Vascular Smooth Muscle α-Actin
Abstract
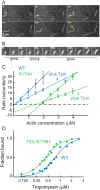
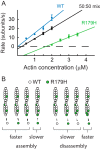
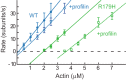
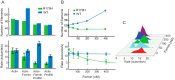
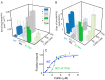
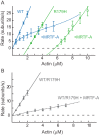
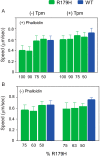
Mutations in vascular smooth muscle α-actin (SM α-actin), encoded by ACTA2, are the most common cause of familial thoracic aortic aneurysms that lead to dissection (TAAD). The R179H mutation has a poor patient prognosis and is unique in causing multisystemic smooth muscle dysfunction (Milewicz, D. M., Østergaard, J. R., Ala-Kokko, L. M., Khan, N., Grange, D. K., Mendoza-Londono, R., Bradley, T. J., Olney, A. H., Ades, L., Maher, J. F., Guo, D., Buja, L. M., Kim, D., Hyland, J. C., and Regalado, E. S. (2010) Am. J. Med. Genet. A 152A, 2437-2443). Here, we characterize this mutation in expressed human SM α-actin. R179H actin shows severe polymerization defects, with a 40-fold higher critical concentration for assembly than WT SM α-actin, driven by a high disassembly rate. The mutant filaments are more readily severed by cofilin. Both defects are attenuated by copolymerization with WT. The R179H monomer binds more tightly to profilin, and formin binding suppresses nucleation and slows polymerization rates. Linear filaments will thus not be readily formed, and cells expressing R179H actin will likely have increased levels of monomeric G-actin. The cotranscription factor myocardin-related transcription factor-A, which affects cellular phenotype, binds R179H actin with less cooperativity than WT actin. Smooth muscle myosin moves R179H filaments more slowly than WT, even when copolymerized with equimolar amounts of WT. The marked decrease in the ability to form filaments may contribute to the poor patient prognosis and explain why R179H disrupts even visceral smooth muscle cell function where the SM α-actin isoform is present in low amounts. The R179H mutation has the potential to affect actin structure and function in both the contractile domain of the cell and the more dynamic cytoskeletal pool of actin, both of which are required for contraction.
Keywords: actin; cofilin; contractile protein; formin; myosin; profilin; vascular smooth muscle cells.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Milewicz D. M., Østergaard J. R., Ala-Kokko L. M., Khan N., Grange D. K., Mendoza-Londono R., Bradley T. J., Olney A. H., Ades L., Maher J. F., Guo D., Buja L. M., Kim D., Hyland J. C., and Regalado E. S. (2010) De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am. J. Med. Genet. A 152A, 2437–2443 - PMC - PubMed
-
- Guo D. C., Pannu H., Tran-Fadulu V., Papke C. L., Yu R. K., Avidan N., Bourgeois S., Estrera A. L., Safi H. J., Sparks E., Amor D., Ades L., McConnell V., Willoughby C. E., Abuelo D., et al. (2007) Mutations in smooth muscle α-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat. Genet. 39, 1488–1493 - PubMed
-
- Guo D. C., Papke C. L., Tran-Fadulu V., Regalado E. S., Avidan N., Johnson R. J., Kim D. H., Pannu H., Willing M. C., Sparks E., Pyeritz R. E., Singh M. N., Dalman R. L., Grotta J. C., Marian A. J., et al. (2009) Mutations in smooth muscle α-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 84, 617–627 - PMC - PubMed
-
- Morisaki H., Akutsu K., Ogino H., Kondo N., Yamanaka I., Tsutsumi Y., Yoshimuta T., Okajima T., Matsuda H., Minatoya K., Sasaki H., Tanaka H., Ishibashi-Ueda H., and Morisaki T. (2009) Mutation of ACTA2 gene as an important cause of familial and nonfamilial nonsyndromatic thoracic aortic aneurysm and/or dissection (TAAD). Hum Mutat 30, 1406–1411 - PubMed
-
- Regalado E. S., Guo D. C., Prakash S., Bensend T. A., Flynn K., Estrera A., Safi H., Liang D., Hyland J., Child A., Arno G., Boileau C., Jondeau G., Braverman A., Moran R., et al. (2015) Aortic disease presentation and outcome associated with ACTA2 mutations. Circ. Cardiovasc. Genet. 8, 457–464 - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

