Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2
- PMID: 27551080
- PMCID: PMC5018801
- DOI: 10.1073/pnas.1611956113
Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2
Abstract
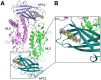
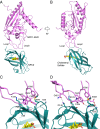
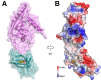
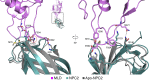
Export of LDL-derived cholesterol from lysosomes requires the cooperation of the integral membrane protein Niemann-Pick C1 (NPC1) and a soluble protein, Niemann-Pick C2 (NPC2). Mutations in the genes encoding these proteins lead to Niemann-Pick disease type C (NPC). NPC2 binds to NPC1's second (middle), lumenally oriented domain (MLD) and transfers cholesterol to NPC1's N-terminal domain (NTD). Here, we report the 2.4-Å resolution crystal structure of a complex of human NPC1-MLD and NPC2 bearing bound cholesterol-3-O-sulfate. NPC1-MLD uses two protruding loops to bind NPC2, analogous to its interaction with the primed Ebola virus glycoprotein. Docking of the NPC1-NPC2 complex onto the full-length NPC1 structure reveals a direct cholesterol transfer tunnel between NPC2 and NTD cholesterol binding pockets, supporting the "hydrophobic hand-off" cholesterol transfer model.
Keywords: Ebola virus glycoprotein; Niemann–Pick type C disease; cholesterol trafficking; crystal structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. - PubMed
-
- Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250(21):8487–8495. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

