MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes
- PMID: 27551152
- PMCID: PMC5030807
- DOI: 10.1084/jem.20160486
MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes
Abstract
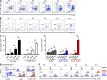
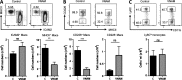
Peritoneal and pleural resident macrophages in the mouse share common features and in each compartment exist as two distinct subpopulations: F4/80(+) macrophages and MHC II(+) CD11c(+) macrophages. F4/80(+) macrophages derive from embryonic precursors, and their maintenance is controlled by Gata6. However, the origin and regulatory factors that maintain MHC II(+) macrophages remain unknown. Here, we show that the MHC II(+) macrophages arise postnatally from CCR2-dependent precursors that resemble monocytes. Monocytes continuously replenish this subset through adulthood. Gene expression analysis identified distinct surface markers like CD226 and revealed that the transcription factor IRF4 was selectively expressed in these macrophages relative to other organs. Monocytes first entered peritoneal or pleural cavities to become MHC II(+) cells that up-regulated CD226 and CD11c later as they continued to mature. In the absence of IRF4 or after administration of oral antibiotics, MHC II(+)CD226(-)CD11c(-) monocyte-derived cells accumulated in peritoneal and pleural cavities, but CD11c(+) CD226(+) macrophages were lost. Thus, MHC II(+) resident peritoneal and pleural macrophages are continuously replenished by blood monocytes recruited to the peritoneal and pleural cavities constitutively, starting after birth, where they require IRF4 and signals likely derived from the microbiome to fully differentiate.
© 2016 Kim et al.
Figures



References
-
- Baldridge M.T., Nice T.J., McCune B.T., Yokoyama C.C., Kambal A., Wheadon M., Diamond M.S., Ivanova Y., Artyomov M., and Virgin H.W.. 2015. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 347:266–269. 10.1126/science.1258025 - DOI - PMC - PubMed
-
- Epelman S., Lavine K.J., Beaudin A.E., Sojka D.K., Carrero J.A., Calderon B., Brija T., Gautier E.L., Ivanov S., Satpathy A.T., et al. . 2014a Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 40:91–104. 10.1016/j.immuni.2013.11.019 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

