Carbapenem Resistance in Acinetobacter baumannii and Other Acinetobacter spp. Causing Neonatal Sepsis: Focus on NDM-1 and Its Linkage to ISAba125
- PMID: 27551277
- PMCID: PMC4976090
- DOI: 10.3389/fmicb.2016.01126
Carbapenem Resistance in Acinetobacter baumannii and Other Acinetobacter spp. Causing Neonatal Sepsis: Focus on NDM-1 and Its Linkage to ISAba125
Abstract
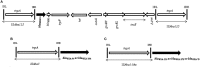
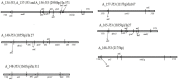
Carbapenem-resistant determinants and their surrounding genetic structure were studied in Acinetobacter spp. from neonatal sepsis cases collected over 7 years at a tertiary care hospital. Acinetobacter spp. (n = 68) were identified by ARDRA followed by susceptibility tests. Oxacillinases, metallo-β-lactamases (MBLs), extended-spectrum β-lactamases and AmpCs, were detected phenotypically and/or by PCR followed by DNA sequencing. Transconjugants possessing the bla NDM-1(New Delhi metallo-β-lactamase) underwent further analysis for plasmids, integrons and associated genes. Genetic environment of the carbapenemases were studied by PCR mapping and DNA sequencing. Multivariate logistic regression was used to identify risk factors for sepsis caused by NDM-1-harboring organisms. A. baumannii (72%) was the predominant species followed by A. calcoaceticus (10%), A. lwoffii (6%), A. nosocomialis (3%), A. junni (3%), A. variabilis (3%), A. haemolyticus (2%), and 14TU (2%). Fifty six percent of the isolates were meropenem-resistant. Oxacillinases present were OXA-23-like, OXA-58-like and OXA-51-like, predominately in A. baumannii. NDM-1 was the dominant MBL (22%) across different Acinetobacter spp. Isolates harboring NDM-1 also possessed bla (VIM-2, PER-1, VEB-2, CTX-M-15), armA, aac(6')Ib, aac(6')Ib-cr genes. bla NDM-1was organized in a composite transposon between two copies of ISAba125 in the isolates irrespective of the species. Further, OXA-23-like gene and OXA-58-like genes were linked with ISAba1 and ISAba3 respectively. Isolates were clonally diverse. Integrons were variable in sequence but not associated with carbapenem resistance. Most commonly found genes in the 5' and 3'conserved segment were aminoglycoside resistance genes (aadB, aadA2, aac4'), non-enzymatic chloramphenicol resistance gene (cmlA1g) and ADP-ribosylation genes (arr2, arr3). Outborn neonates had a significantly higher incidence of sepsis due to NDM-1 harboring isolates than their inborn counterparts. This study demonstrates the significance of both A. baumannii and other species of Acinetobacter in cases of neonatal sepsis over an extended period. Oxacillinases and bla NDM-1 are the major contributors to carbapenem resistance. The dissemination of the bla NDM-1 is likely linked to Tn125 in diverse clones of the isolates.
Keywords: 5′–3′ CS; ARDRA; Acinetobacter spp.; India; NDM-1; OXA-23; Tn125; neonatal sepsis.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

