Base Excision Repair of N6-Deoxyadenosine Adducts of 1,3-Butadiene
- PMID: 27552084
- PMCID: PMC5098428
- DOI: 10.1021/acs.biochem.6b00553
Base Excision Repair of N6-Deoxyadenosine Adducts of 1,3-Butadiene
Abstract
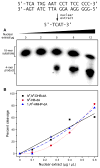
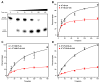
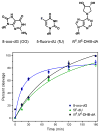
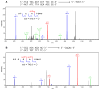
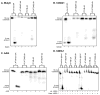
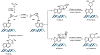
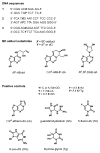
The important industrial and environmental carcinogen 1,3-butadiene (BD) forms a range of adenine adducts in DNA, including N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine (N6-HB-dA), 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine (1,N6-HMHP-dA), and N6,N6-(2,3-dihydroxybutan-1,4-diyl)-2'-deoxyadenosine (N6,N6-DHB-dA). If not removed prior to DNA replication, these lesions can contribute to A → T and A → G mutations commonly observed following exposure to BD and its metabolites. In this study, base excision repair of BD-induced 2'-deoxyadenosine (BD-dA) lesions was investigated. Synthetic DNA duplexes containing site-specific and stereospecific (S)-N6-HB-dA, (R,S)-1,N6-HMHP-dA, and (R,R)-N6,N6-DHB-dA adducts were prepared by a postoligomerization strategy. Incision assays with nuclear extracts from human fibrosarcoma (HT1080) cells have revealed that BD-dA adducts were recognized and cleaved by a BER mechanism, with the relative excision efficiency decreasing in the following order: (S)-N6-HB-dA > (R,R)-N6,N6-DHB-dA > (R,S)-1,N6-HMHP-dA. The extent of strand cleavage at the adduct site was decreased in the presence of BER inhibitor methoxyamine and by competitor duplexes containing known BER substrates. Similar strand cleavage assays conducted using several eukaryotic DNA glycosylases/lyases (AAG, Mutyh, hNEIL1, and hOGG1) have failed to observe correct incision products at the BD-dA lesion sites, suggesting that a different BER enzyme may be involved in the removal of BD-dA adducts in human cells.
Figures


References
-
- Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. - PubMed
-
- White WC. Butadiene production process overview. Chem Biol Interact. 2007;166:10–14. - PubMed
-
- Hoffmann D, Hoffmann I, el Bayoumy K. The less harmful cigarette: a controversial issue a tribute to Ernst L Wynder. Chem Res Toxicol. 2001;14:767–790. - PubMed
-
- Rice JM, Boffetta P. 1,3-Butadiene, isoprene and chloroprene: reviews by the IARC monographs programme, outstanding issues, and research priorities in epidemiology. Chem Biol Interact. 2001;135–136:11–26. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

